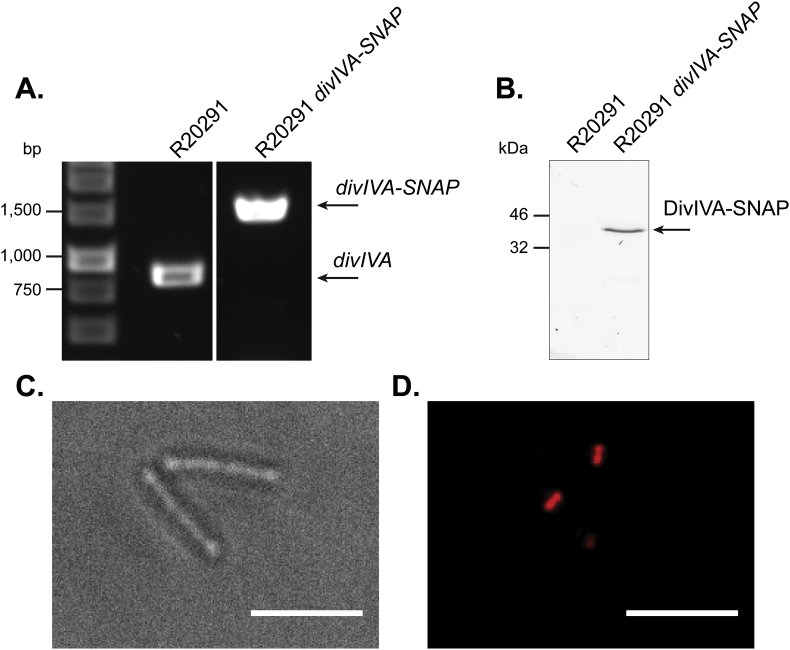
Fig. 3.
Precise manipulation of the R20291 genome accelerated by the use of optimised conjugation. (A) 0.8% Agarose gel showing PCR fragments amplified using primers flanking the SNAP-tag coding sequence insertion site in the chromosomal divIVA gene. The increase of approximately 550bp from 885 bp (R20291) to 1443 bp (R20291 divIVA-SNAP) suggests the correct insertion of the SNAP-tag coding sequence. (B) 12% SDS-PAGE gel imaged using a fluorescence imager, showing resolved lysates from SNAP TMR-star treated R20291 and R20291divIVA-SNAP. A band at approximately 40 kDa in the mutant corresponds to the addition of a SNAP-tag on DivIVA. (C) Brightfield and (D) fluorescence microscopy of exponentially growing R20291divIVA-SNAP stained with 250 nM SNAP TMR-star for 30 min, showing mostly septal and some polar localisation of fluorescence. This demonstrates successful modification of the R20291 genome. Scale bar represents 5 μm.