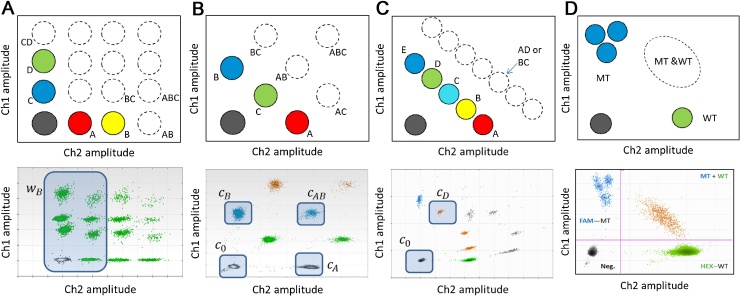
Fig. 2.
Graphical outputs for higher order multiplexing strategies. All illustrative examples given here used the QX200™ Droplet Digital™ PCR System (Bio-Rad). For each multiplexing strategy a schematic and worked example of the configuration of clusters in the 2D plot is given with the amplitude in channel 1 (ch1) is represented on the y-axis with the amplitude in channel 2 (ch2) is represented on the x-axis. For each schematic the approximate location of the single target positive clusters are identified as solid coloured circles with the double or triple target positive clusters are shown as dotted circles. For the worked examples, the clusters are not pseudocoloured according to the target due to the limitations of the software. (A) For amplitude-based multiplexing, four independent targets are being quantified: targets A (100% Ch2-labelled probe), B (100% Ch2), C (50% Ch1) and D (100% Ch1) giving 16 possible clusters in the 2D amplitude space. For the worked example, all the clusters containing partitions that do not contain target B are lassoed (wB). (B) For ratio-based multiplexing, three independent targets are being quantified: targets A (100% Ch2), B (100% Ch1) and C (50% Ch2 and 50% Ch1) giving 8 possible clusters in the 2D amplitude space. For the worked example, the four clusters used in the quantification are lassoed and labelled: negative cluster for all targets (c0), single-positive for target B (cB), single-positive for target A (cA), and double-positive for targets A and B (cAB). (C) For ratio-based non-discriminating multiplexing, five targets are being quantified: targets A (100% Ch2), B (75% Ch2/25%Ch1), C (50% Ch2/50% Ch1), D (25% Ch2/75% Ch1) and E (100% Ch1). A total of 32 combinations of clusters are possible, however, due to the ratios of the probes for the different targets, most are not uniquely identifiable. For example, a cluster containing both targets A and D (125% Ch2/75% Ch1) would not be distinguishable from a cluster containing both targets B and C (also 125% Ch2/75% Ch1). Therefore only the single-positive clusters can be used in the quantification. For the worked example, the two clusters used for the quantification of target D are lassoed: negative cluster for all targets (c0) and single-positive cluster for target D (cD). (D) For non-discriminating multiplexing the example of the KRAS mutations in codons 12 and 13 is shown. The 7 probes for the mutant SNVs are all conjugated with the Ch1-labelled probe and the WT probe is conjugated with the Ch2-labelled probe. Three diffuse clusters are visible in the mutant probe channel with a single cluster in the WT channel. The double-positive cluster is also visible. It is not possible to discriminate between the 3 mutant clusters and so this set up allows quantification of the total number of mutant sequences only.