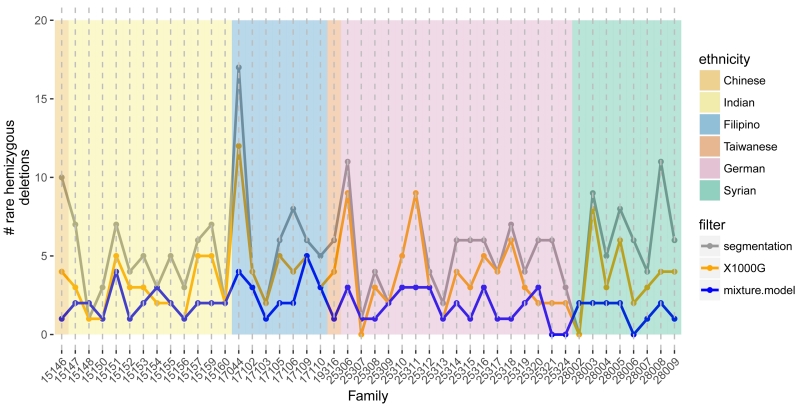
Figure 1.
The number of autosomal hemizygous deletions (y-axis) identified among 95 participants across 46 mulitiplex families (x-axis). Candidate deletions were first identified by segmentation of M values (gray). Excluding deletions overlapping with homozygous deletions and copy number polymorphisms in the 1000G project, we obtained an initial estimate of the frequency of rare, autosomal hemizygous deletions per family (orange). At each region with a potentially rare deletion, we fit Bayesian mixture models with and without a mixture component for the hemizygous copy number state to the average M values. For regions where the log Bayes factor comparing the model with deletion to the model without deletion was at least 2, a sample was considered hemizygous if the posterior probability for the hemizygous component was at least 0.9. Excluding regions with more than 5 families identified as hemizygous under this mixture model, a total of 88 rare deletions were identified in the 46 families with a median frequency per family of 2 (blue).