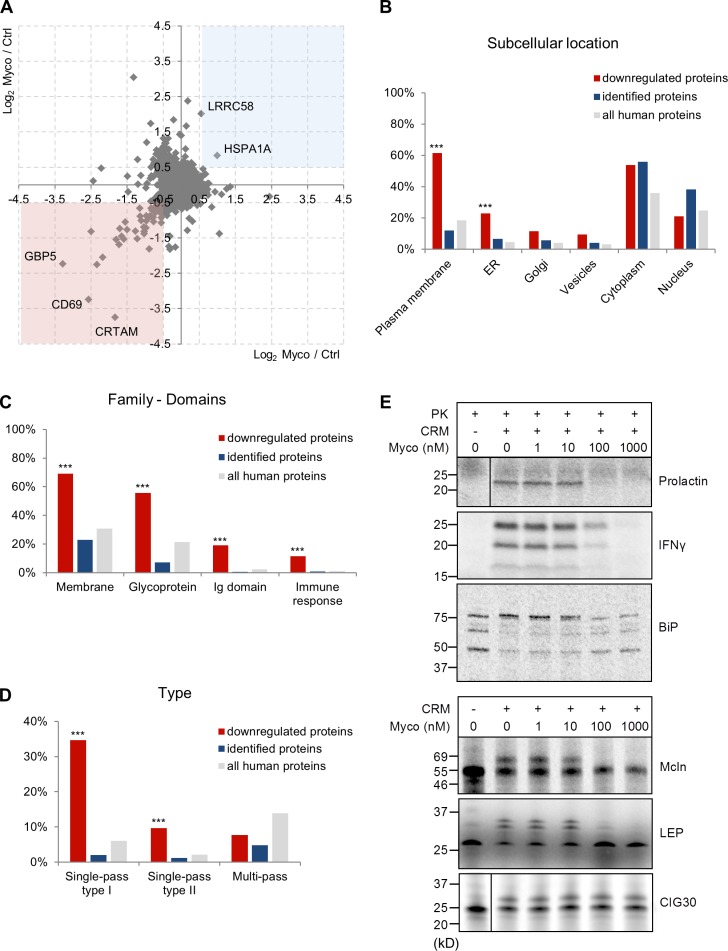
Figure 2.
Mycolactone is a broad-spectrum inhibitor of Sec61. (A) Scatter plot showing the log2 SILAC ratios for individual proteins quantified in analysis 1 on the x axis (light condition: 40 nM mycolactone [Myco]; heavy condition: vehicle control [Ctrl]) and analysis 2 on the y axis (reversed conditions). Proteins with a log2 ratio <−0.5 (pink square) or >0.5 (blue square) in both analyses were considered modulated by mycolactone. CRTAM, cytotoxic and regulatory T cell molecule. (B and C) Gene ontology cellular component (B) and SwissProt Protein Information resource keywords (C) annotation analyses of the proteins that were reproducibly down-regulated in mycolactone-exposed T cells (red; n = 52), compared with identified proteins (blue; n = 6,503) and all human proteins in UniProt (gray; n = 20,204). (D) Distribution of downregulated (red), identified (blue), and all human proteins (gray) over different categories of membrane proteins. (B–D) Statistics were calculated by Fisher exact tests comparing downregulated versus identified proteins. ***, P < 0.001. (E) IVT assays of various Sec61 clients in the presence of increasing concentrations of mycolactone. ER translocation of nonglycosylated proteins (prolactin, IFN-γ, and BiP) was assessed by treatment with proteinase K (PK), with resistance to proteinase K indicating correct translocation into the ER lumen. Detergent-treated controls are shown in Fig. S1 C. It should be noted that BiP is largely protease resistant and, upon proteinase K treatment, forms shorter fragments. Translocation of glycosylated proteins (Mcln, LEP, and CIG30) was assessed by analyzing the change in migration in SDS-PAGE and autoradiography. Glycosidase-treated controls are shown in Fig. S1 C. The data shown are from one of two independent experiments, which gave similar results.