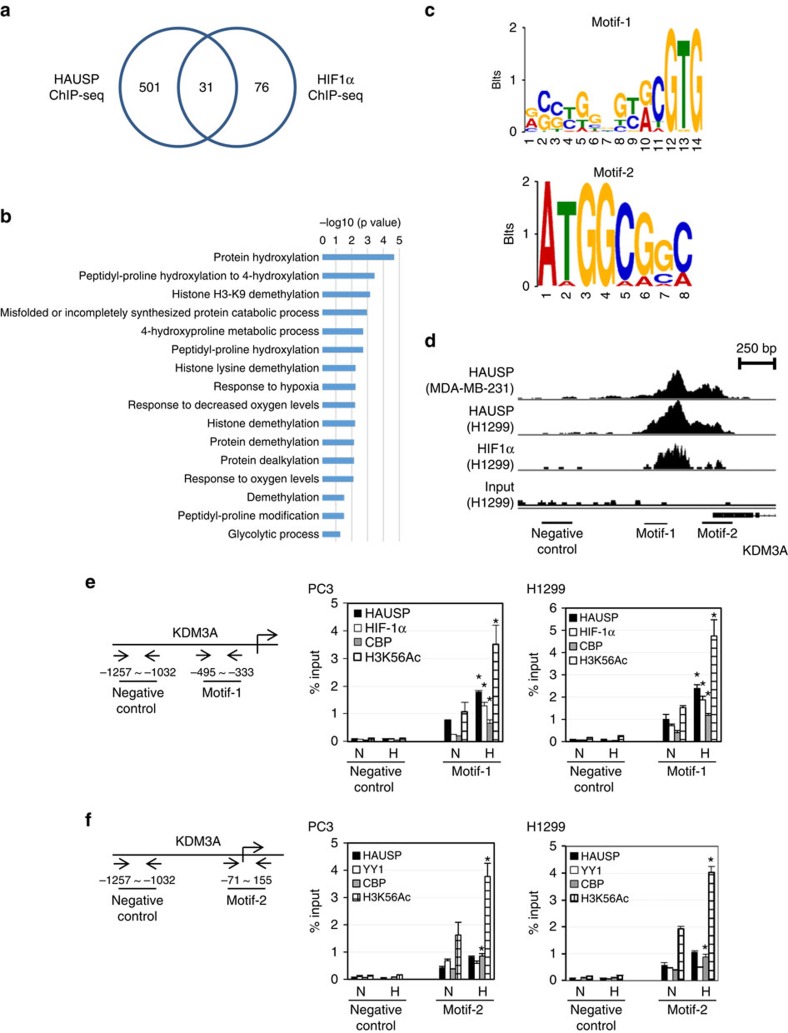
Figure 6. Analysis of ChIP-seq results with identification of two motifs and confirmation of various factors binding to the KDM3A promoter regions.
(a) The numbers of overlapping set of genes that were bound by HAUSP and HIF-1α by ChIP-seq analysis. (b) GO analysis of the 31 genes that showed binding by both HAUSP and HIF-1α. (c) Identification of two consensus motifs that were bound by HAUSP and HIF-1α. (d) Gene track analysis of a representative gene, KDM3A, that contained motif-1 and motif-2 in its promoter regions. (e) The left panel shows the schematic diagram of the KDM3A promoter regions containing motif-1 that were checked by qChIP assays. qChIP analysis confirmed the binding of various factors to motif-1 in the KDM3A promoter regions. (f) The left panel shows the schematic diagram of the KDM3A promoter regions containing motif-2 that were checked by qChIP assays. qChIP analysis confirmed the binding of various factors to motif-2 in the KDM3A promoter regions. N, normoxia; H, hypoxia. Data from three independent experiments are expressed as mean±s.d. The asterisk (*) indicated statistical significance (P<0.05) between normoxia and hypoxia conditions, t-test.