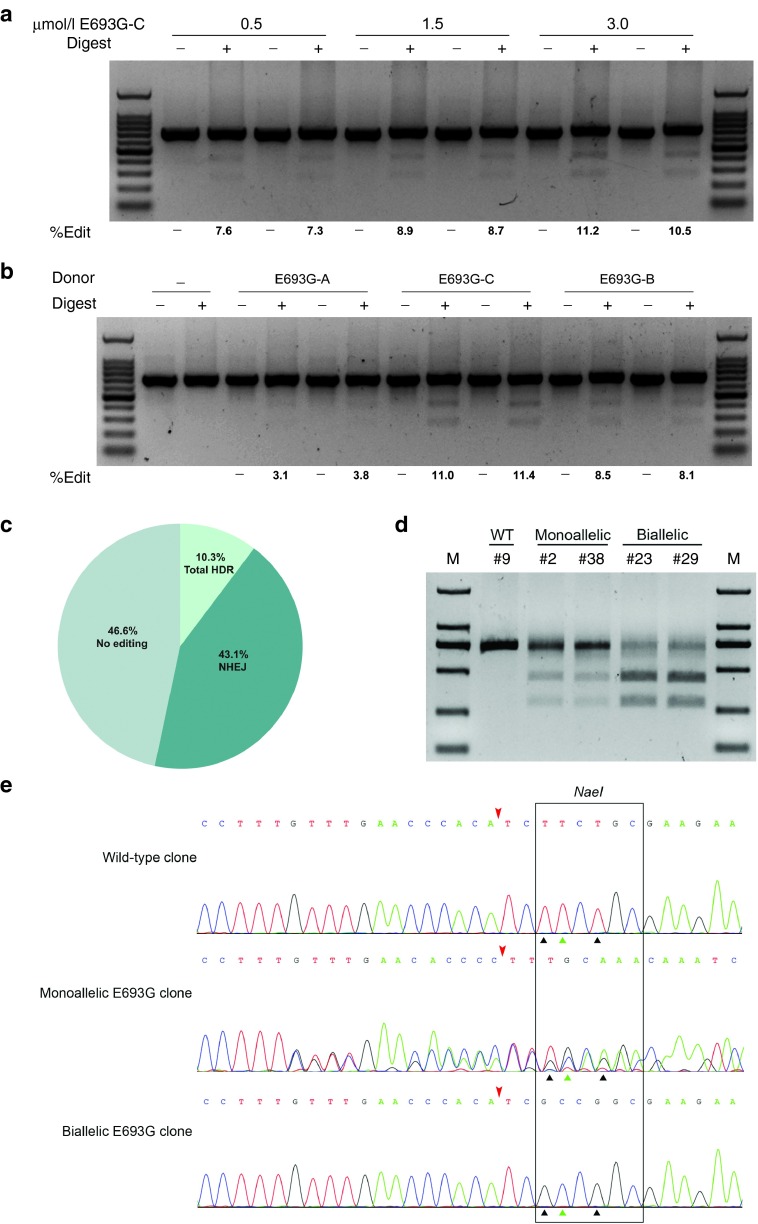
Figure 2.
CRISPR/Cas9-based ssODN-mediated knockin of point mutations at porcine APP locus. (a) The 120-nt ssODN donor was transfected into porcine fetal fibroblasts (PFFs) with different doses to determine the optimal dosage for homology-directed repair (HDR). (b) Four ssODN donors with different lengths of homology arms were transfected into PFFs. The level of HDR was analyzed by the RFLP assay and is shown at the bottom of each gel. The electroporation was performed in biological duplicate. (c) PFF colonies derived from one single cell were isolated by the limiting dilution method. The mutation patterns (NHEJ, HDR, or no editing) for all colonies were shown. (d) RFLP analysis of colonies harboring monoallelic or biallelic HDR alleles. Purified PCR products encompassing the target regions were digested with NaeI. (e) Sequencing chromatogram of representative HDR colonies. The Cas9 cleavage sites were marked with red arrowheads. The green triangles indicate the mutation for p.E693G, while black triangles indicate the synonymous mutations.