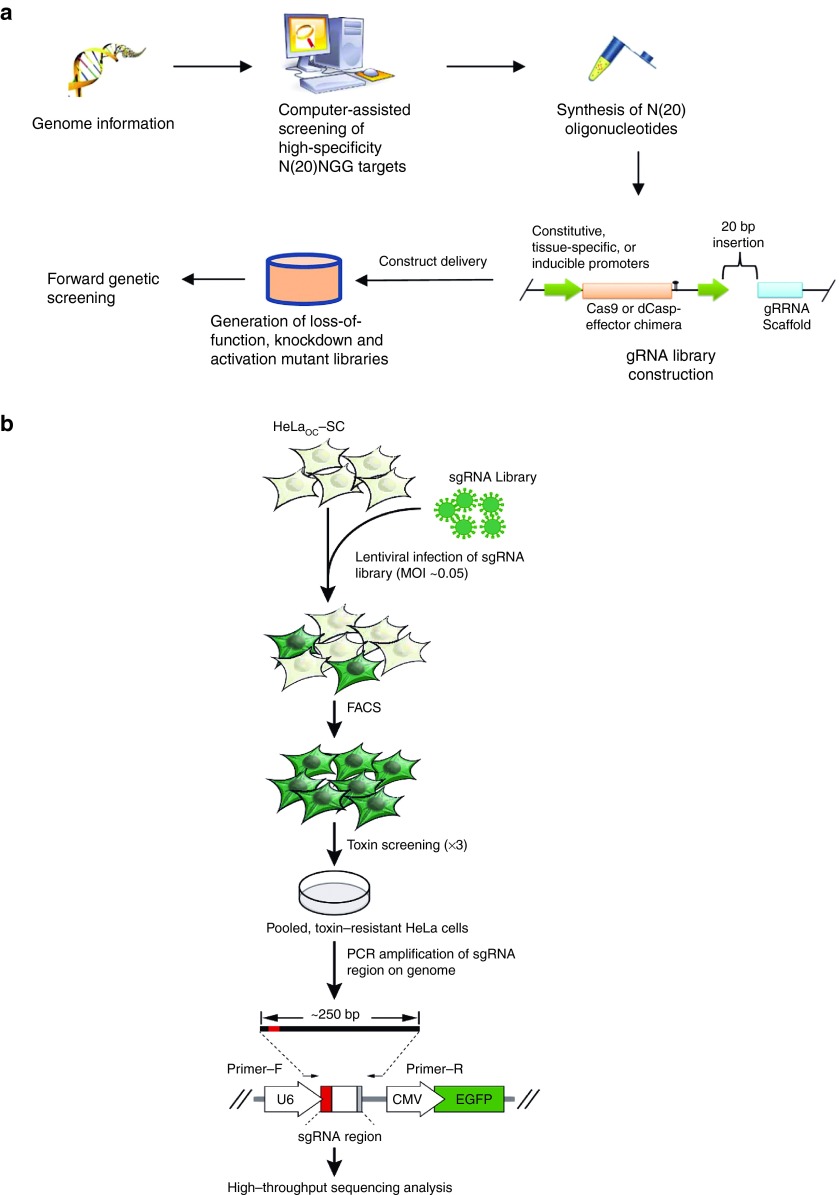
Figure 5.
Flowchart for Cas9-based high-throughput forward genetic screening. Reasonable and precise N20NGG targetable areas on the other side of the complete genome were chosen using computer-assisted programs. DNA oligonucleotides with N20 sequences were synthesized as well as adjusted for ligation with a construct, which produced a gRNA library that can coexpress Cas9 or a Cas9-effector chimera and varied gRNAs propelled by constitutive, tissue-specific, or inducible promoters (a).26 The gRNA library was then transported to host cells to produce varied mutant libraries for forward genetic screening (b).43 sgRNAs were transported into HeLaOC-SC cells using lentiviral infection with a MOI of 0.05. Library cells steadily expressing sgRNAs were gathered using FACS for green fluorescence 48 hours postinfection. Library screening was performed with three toxin courses, followed by PCR amplification of the sgRNA-coding sequence incorporated into the chromosomes. The purified PCR product then underwent high-throughput sequencing analysis using Illumina's HiSeq 2500.