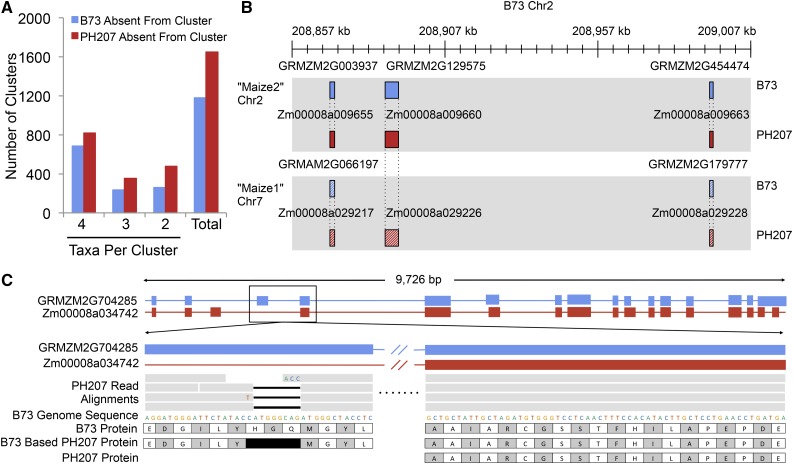
Figure 7.
Lessons Learned from Comparisons between Genome Assemblies of Elite Maize Inbred Lines.
(A) Homologous gene clusters between Brachypodium, maize B73, maize PH207, rice, and S. bicolor that contain a maize gene from only B73 or PH207 and at least one other species.
(B) Example of a duplicated gene showing differential fractionation between B73 and PH207. Maize is an ancient tetraploid that has undergone genome fractionation. A previous study identified and classified genes in the B73 genome into two maize subgenomes, Maize1 and Maize2 (Schnable et al., 2011). The middle genes in these syntenic blocks are showing differential fractionation between B73 and PH207, with PH207 retaining both copies and B73 losing the copy in the Maize2 subgenome (gene positions: GRMZM2G003937 chr2:208,869,363..208,870,921; GRMZM2G129575 chr2:208887428..208891800; GRMZM2G454474 chr2:208,993,547..208,994,705; Zm00008a009655 chr2:212,248,102..212,249,027; Zm00008a009660 chr2:212,373,566..212,385,564; Zm00008a009663 chr2: 212,427,728..212,428,519; GRMZM2G066197 chr7:161,658,677..161,660,464; GRMZM2G179777 chr7:161,843,771..161,845,175; Zm00008a029217 chr7:161,948,327..161,949,627; Zm00008a029226 chr7:162,105,478..162,112,256; Zm00008a029228 chr7:162,164,382..162,166,725).
(C) Example of a gene containing a putative frameshift that is compensated by alternative intron/exon boundaries when using annotation specific to the individual. PH207 reads were aligned to the B73 reference genome assembly and putative large effect variants were identified based on the B73 annotation. The putative frameshift variant shown in this figure is corrected for by an alternative gene model in PH207 that was identified using PH207-specific evidence to annotate genes in the PH207 genome assembly.