Figure 8.
The Upstream Conserved Sequence Affects B-Dependent mRNA Degradation.
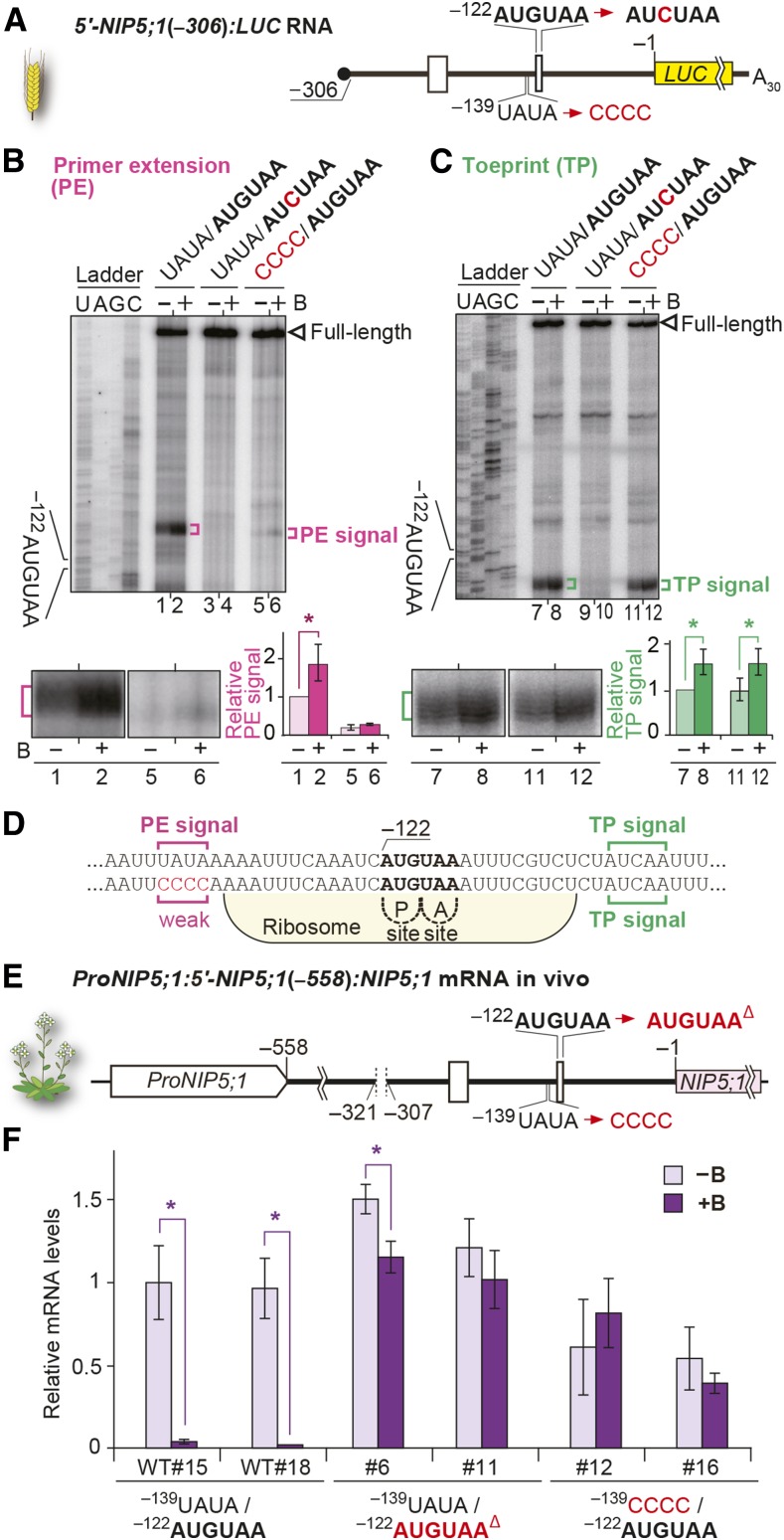
(A) Schematic representation of 5′-NIP5;1(−306):LUC RNA. Open boxes represent uORFs.
(B) and (C) RNA carrying 5′-NIP5;1(−306):LUC with or without a mutation in AUGUAA or with a mutation in the region 17 to 14 nucleotides upstream of −122AUG-stop was translated in WGE. Primer extension (B) and toeprint (C) analyses in WGE with 300 µM B (+B) or without B supplementation (−B). Signals are enlarged under the main image of the AUGUAA lane, and their relative intensities are shown. Means ± sd are shown (n = 3). Asterisks indicate significant differences (P < 0.05).
(D) Nucleotide sequence around −122AUG-stop. Positions of primer extension (magenta) and toeprint signals (green) are marked. Ribosome occupation of RNA having the AUG codon positioned at the P-site is shown.
(E) Schematic representation of ProNIP5;1:5′-NIP5;1(−558):NIP5;1. Deletion of −312AUGUAA, a mutation in the upstream conserved sequence, and the deletion of −122AUGUAA are indicated.
(F) mRNA accumulation in transgenic plant roots grown under 100 µM B (+B) and 0.3 µM B (−B) conditions. Means ± sd of relative mRNA accumulation (n = 3) are shown. Asterisks indicate significant reductions under +B condition (P < 0.05). Two independently transformed lines were used for each construct.