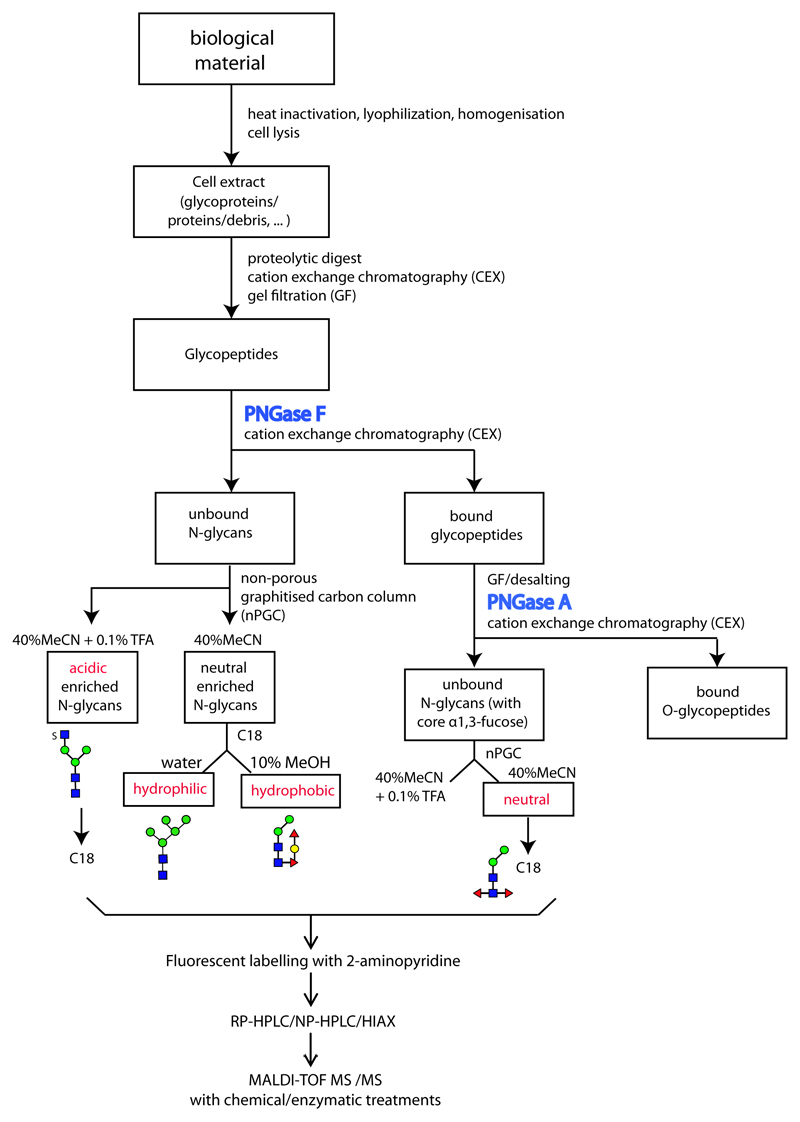
Figure 1. Glycomic workflow for the analysis of N-glycans.
Initially, samples are proteolysed, the glycopeptides enriched by cation exchange (CEX) and gel filtration (GF) and the glycans released enzymatically, whereby PNGase A (and not PNGase F) is capable of releasing the core α1,3-fucosylated N-glycans. Subsequent sub-fractionation by non-porous graphitised carbon (NPGC) and/or reversed-phase (C18) resins result in pools differing in terms of anionic and zwitterionic modifications. Finally, all N-glycans are analysed by different types of HPLC (reversed or normal phase or hydrophilic interaction/anion exchange; RP, NP or HIAX) in combination with MALDI-TOF MS/MS and chemical/enzymatic treatments. The remaining O-glycopeptides can be subject to β-elimination and LC-MS. The example structures, shown according to the nomenclature of the Consortium for Functional Glycomics, are from a marine snail.(12)