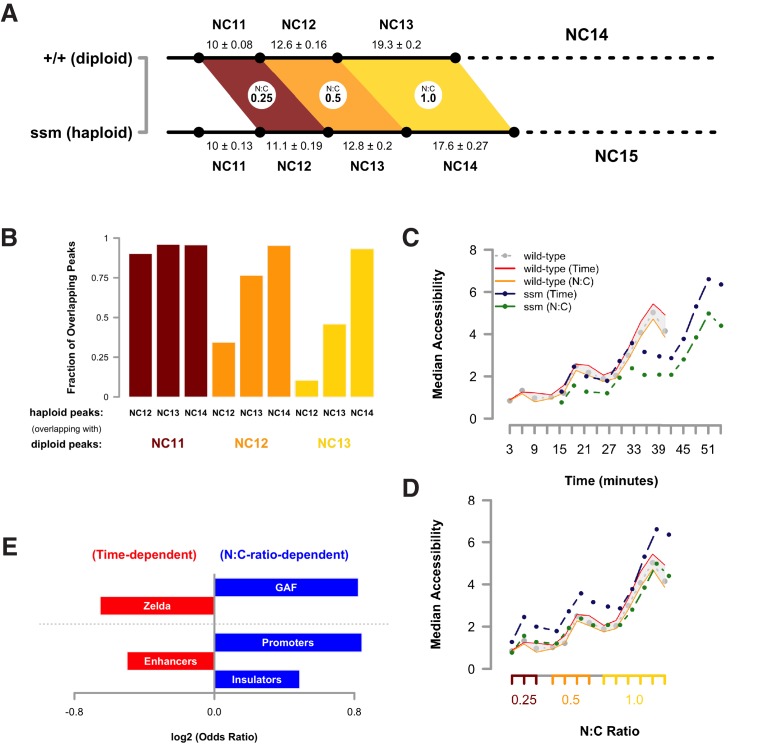
Figure 2. Dynamic acquisition of chromatin accessibility is regulated by the N:C ratio.
(A) Cell cycle times for diploid (+/+, n = 21) and haploid embryos (ssm, n = 12) were measured by time lapse confocal imaging of His2Av-GFP. Mean cell cycle times ± SEM are indicated. Colored regions indicate periods of equivalent N:C ratios (0.25–1.0) between genotypes. (B) Accessible peaks were identified for haploid embryos and the fraction of overlap or co-occurence between diploids and haploids was calculated and plotted for the indicated cell cycles. (C) Median accessibility for haploid and diploid embryos was calculated for the sets of regions indicated in the legend (n = 2630 N:C ratio dependent regions, n = 1658 time-dependent regions). All differences in magnitude between time- and N:C ratio-dependent peaks in haploid embryos are statistically significant at p<0.01 by a randomization test, whereas only the NC13 + 15’ time point achieves p<0.01 in the diploid dataset. Significance testing performed by proportionally dividing all datapoints into two groups at random and testing whether observed differences in median accessibility were equal to or greater than the difference observed between the ‘N:C ratio’ and ‘time’ groupings (one-tailed permutation test for n = 1×104 trials). (D) Median accessibility data from panel C was re-scaled along the x-axis to match relative N:C ratios as shown in panel A. (E) Odds ratios for enrichment of the indicated genomic features and transcriptional regulators were calculated. Time-dependent chromatin accessibility is enriched for enhancers (p=5.78x10−7) and Zelda binding (p=2.42x10−12), whereas N:C ratio dependent chromatin accessibility is enriched for promoters (p=5.66x10−17), insulators (p=2.42x10−4), and GAF binding (p=2.72x10−09). p-Values are from two-sided Fisher’s exact test on contingency tables constructed on [-/+ feature by time/N:C]. GAF odds ratios were calculated from regions that were GAF+ and Zelda-. GAF is significantly enriched in both N:C and time-dependent classes if co-occurrence with Zelda is considered.