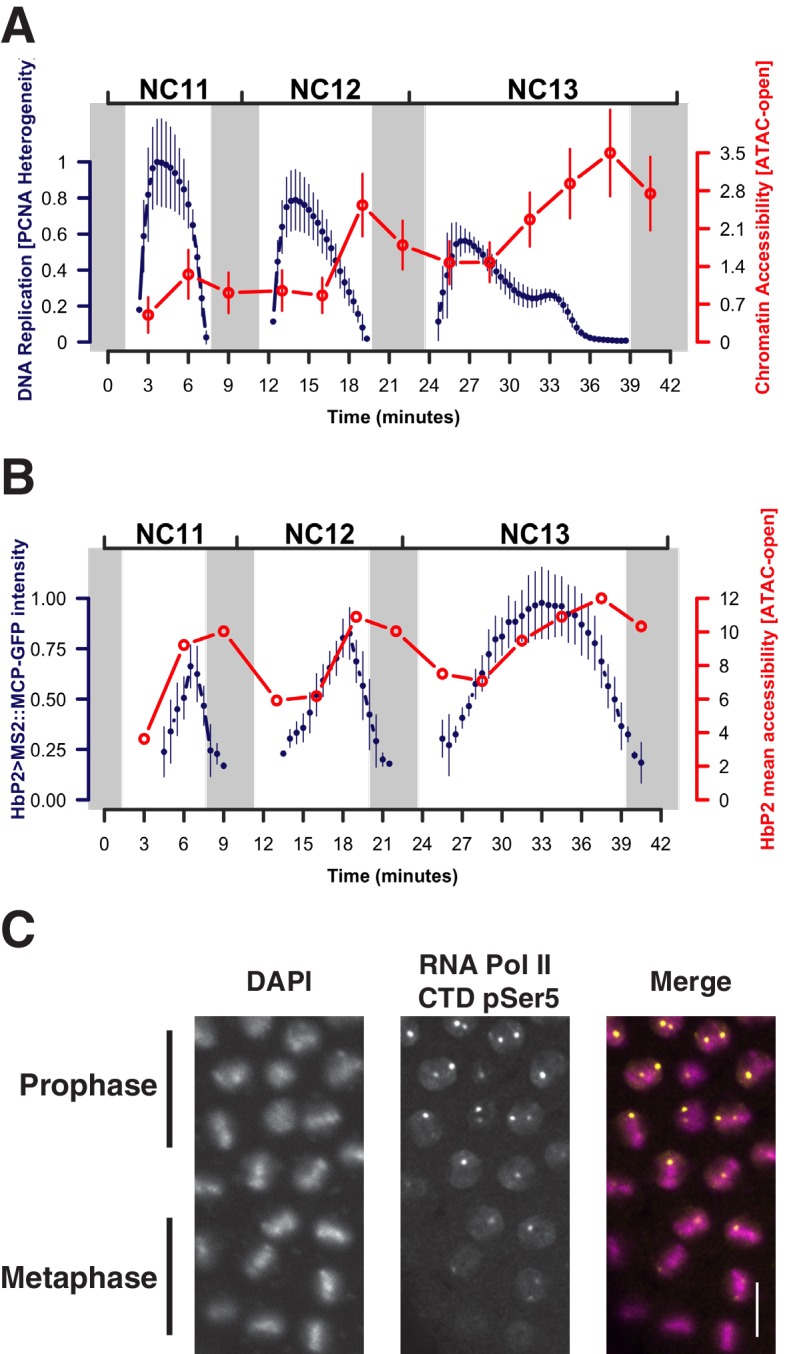
Figure 3. Patterns of stable and dynamic chromatin accessibility over the cell cycle.
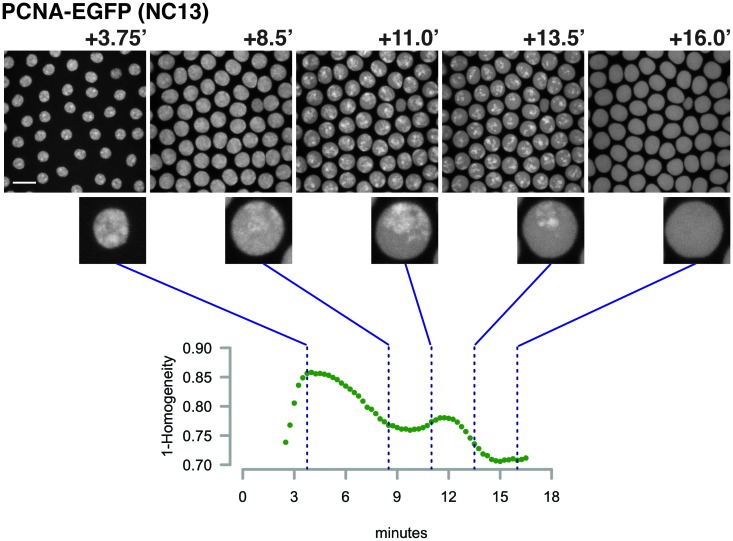
(A) Genome-wide mean ATAC-open coverage is plotted (red, error bars show std. dev. between peaks, n = 9824) over the mean scaled heterogeneity in PCNA-EGFP to estimate DNA replication activity (blue, n = 4, error bars show std. dev. between embryos). Mitotic phases are indicated by grey shading. (B) Mean coverage of sequencing reads from accessible chromatin over the hunchback P2 promoter/enhancer is plotted (red) over the mean measured spot fluorescence intensity from a hunchback P2> MS2(24) LacZ: : MCP-GFP reporter (blue, n = 3, error bars show std. dev. between embryos). Mitotic phases are indicated by grey shading. (C) Immunofluorecence for chromatin morphology (DAPI) and RNA Polymerase II (CTD pSer5) in a region of an NC13 embryo transitioning from prophase to metaphase is shown. Observed mitotic states are indicated at left. Scale bar = 10 μm.