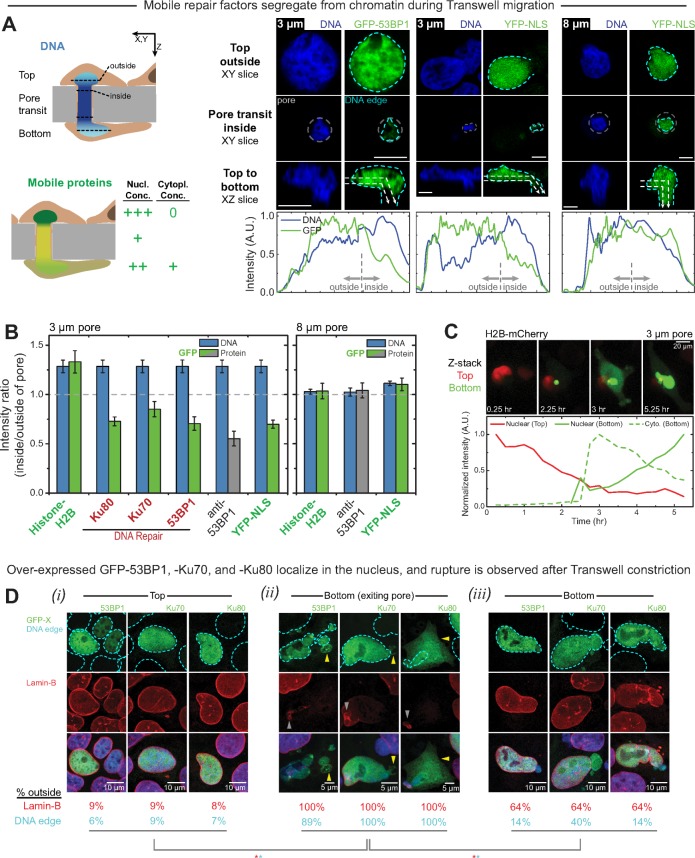
FIGURE 1:
Migration through small pores compacts the chromatin and causes local depletion of mobile nuclear proteins plus occasional nuclear rupture. (A) Schematics illustrate that as a cell nucleus squeezes into a small pore, its chromatin becomes compact, and the opposite occurs with mobile proteins. Confocal sections of fixed cells show the same as a nucleus squeezes into a 3-μm pore, with mobile proteins such as GFP-53BP1 and YFP-NLS-MS2 decreasing in density within the small pore (representative of ≥20 cells/protein). Pore exits show similar differences. Large pores of 8 μm show no such DNA compaction or protein depletion. Intensity profiles for DNA and protein are each normalized to the highest values along the dashed arrows shown in the XZ slices (bar, 5 μm). (B) Segregation of mobile proteins, including endogenous 53BP1, away from chromatin is evident at both the entrance and exit of 3-μm pores. Because DNA is enriched inside the pore, its intensity ratio tends above 1; the same applies for histone-H2B. Conversely, mobile proteins accumulate outside the pore, so their intensity ratios fall consistently below 1. Segregation does not occur in 8-μm pores (≥20 cells/group, at least three experiments). (C) Live imaging of H2B-mCherry–overexpressing U2OS cells reveals nuclear rupture—with leakage of H2B into the cytoplasm—followed by nuclear relocalization over hours (representative of at least three experiments). (D) At the top and bottom of a Transwell membrane, GFP-53BP1, -Ku70, and -Ku80 localize in the nucleus. For cells exiting 3-μm pores, rupture is sometimes evident with mobile proteins in the cytoplasm based on GFP proteins outside the boundary of lamin-B (red) and/or the DNA edge (blue) (≥9 exiting cells/group, ≥100 top cells/group; *p < 0.05). The percentage of cells with mislocalized GFP protein is likewise significantly greater for exiting cells than for cells that fully migrated to the bottom (≥35 bottom cells/group; *p < 0.05).