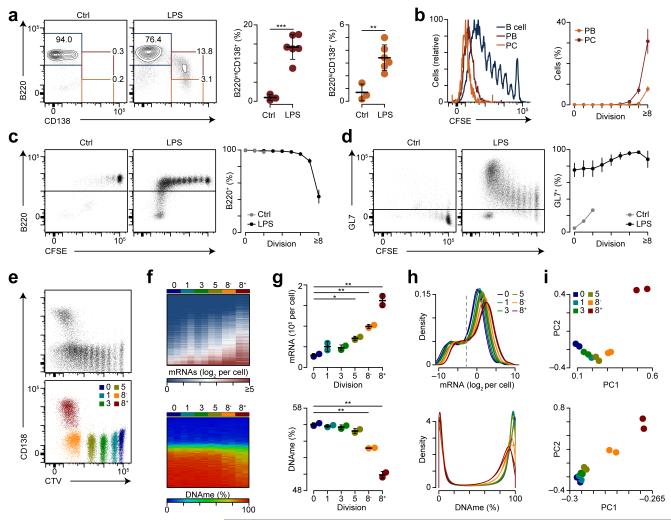
Figure 5.
Transcriptional amplification and DNA hypomethylation coincide with cellular division. (a) B220 and CD138 expression on transferred splenic naïve CD45.1+B220+ B cells showing LPS-induced CD138 expression (left). Quantification of B220intCD138+ plasmablasts (PBs; center) and B220loCD138+ plasma cells (PCs; right). (b) CFSE dilution on B220+ B cells, B220intCD138+ plasmablasts, and B220loCD138+ plasma cells (left). Quantification of B220intCD138+ plasmablasts and B220loCD138+ plasma cells by division (right). (c) B220 expression by division for control and LPS challenged mice (left). Quantitation of B220+ cells by division (right). (d) GL7 expression by division for control and LPS challenged mice (left). Quantitation of GL7+ cells by division (right). (e) Cell trace violet (CTV) staining and CD138 expression for CD45.1+ cells (top) and post sort purity (bottom) for the indicated populations. (f) Heatmaps of transcript expression (top) and DNA methylation (DNAme) (bottom) for populations shown in e. (g) Quantification of average mRNAs per cell (top) and DNAme (bottom) measured across 1,639,598 CpGs assessed. (h) Probability density for mRNA expression (top) and DNAme (bottom). The dashed gray line indicates the 90% detection level. (i) Principle component analysis for mRNA expression (top) and DNAme (bottom). *P <0.05, **P <0.01 and ***P <0.001 (two-tailed t-test (a, g)). Data are from two experiments with 4 and 6 mice per experiment (a-e) or one experiment performed in biological duplicate (f-i) (a-d, g; mean and s.d.).