Abstract
Bluetongue is an economically important infectious, arthropod borne viral disease of domestic and wild ruminants, caused by Bluetongue virus (BTV). Sheep are considered the most susceptible hosts, while cattle, buffalo and goats serve as reservoirs. The viral pathogenesis of BTV resulting in presence or absence of clinical disease among different hosts is not clearly understood. In the present study, transcriptome of sheep and goats peripheral blood mononuclear cells infected with BTV-16 was explored. The differentially expressed genes (DEGs) identified were found to be significantly enriched for immune system processes - NFκB signaling, MAPK signaling, Ras signaling, NOD signaling, RIG signaling, TNF signaling, TLR signaling, JAK-STAT signaling and VEGF signaling pathways. Greater numbers of DEGs were found to be involved in immune system processes in goats than in sheep. Interestingly, the DEHC (differentially expressed highly connected) gene network was found to be dense in goats than in sheep. Majority of the DEHC genes in the network were upregulated in goats but down-regulated in sheep. The network of differentially expressed immune genes with the other genes further confirmed these findings. Interferon stimulated genes - IFIT1 (ISG56), IFIT2 (ISG54) and IFIT3 (ISG60) responsible for antiviral state in the host were found to be upregulated in both the species. STAT2 was the TF commonly identified to co-regulate the DEGs, with its network showing genes that are downregulated in sheep but upregulated in goats. The genes dysregulated and the networks perturbed in the present study indicate host variability with a positive shift in immune response to BTV in goats than in sheep.
Keywords: Bluetongue, Bluetongue virus serotype-16, PBMCs, transcriptome, RNA-seq, DEGs, DEHC genes
1. Introduction
Bluetongue (BT) is an economically important infectious, arthropod-borne viral disease of sheep, cattle, goats, and wild ruminants, caused by Bluetongue virus (BTV).·Cattle are an important reservoir for sheep and other susceptible ruminants; some wild ruminant species also may be reservoirs. It is characterized by rise of temperature (104–106 °F) along with salivation and frothing, swelling of tongue and lips, petechial hemorrhages in oral mucous membranes and inflammatory reaction on coronary bands resulting in decreased production [1]. BT causes severe economic losses in terms of morbidity, mortality, production, and reproduction and animal related trade restrictions [2]. It is transmitted by biting midges of the genus Culicoides, family Ceratopogonidae. Thus, considering its economic impact on the small ruminants, World Organization for Animal Health (formerly the Office International des Epizooties (OIE)) listed Bluetongue as a “notifiable disease”. It is among the main transboundary diseases affecting small ruminant population in developing countries.
BTV the prototype member of the genus Orbivirus, family Reoviridae is a non-enveloped, icosahedral, double stranded RNA virus having ten segments in the genome that encode seven structural (VP1 to VP7) and four non-structural proteins (NS1, NS2, NS3 NS3a) [3], [4], [5]. Recently, two more non-structural proteins (NS4 and NS5) have been reported [6], [7]. The BTV core is composed of two major proteins (VP3 and VP7) and three minor proteins (VP1, VP4 and VP6). Outer capsid (VP2 and VP5) surrounds the core, but is removed during cell entry. A total of 27 serotypes of BTV have been described in the world [8], [9], [10], [11]. Out of these, 22 serotypes have been reported in India. The emergence of new serotypes is mainly attributed to mutations and genetic reassortments of segmented genome of BTV [12].
BTV was found to replicate within mononuclear phagocytic and endothelial cells, lymphocytes and possibly other cell types in lymphoid tissues, the lungs, skin and other tissues [13]. Infected ruminants may exhibit a prolonged viraemia following which the virus could be identified in a number of tissues and organs. BTV replication in agranular leucocytes that are morphologically similar to lymphocytes, monocytes and dendritic cells or macrophages has been confirmed using confocal microscopy [14]. Monocytes infected with BTV in vivo showed cytopathic effect (CPE) after a few days of post-infection, while freshly isolated lymphocytes were resistant to infection [15], [16]. The fate of BTV-infected lymphocytes is still poorly understood, and there is experimental evidence suggesting that different subsets can be lytically or non-lytically infected [15], [17]. Overall, it seems likely that several leucocyte subsets can support BTV replication and may be involved in viral dissemination in the ruminant host. However, currently it is still unclear whether infected leucocytes are lytically or non-lytically infected or whether they are eliminated by the immune system in vivo. Also, the viral pathogenesis of BTV resulting in presence or absence of clinical disease among different hosts is not clearly understood and both, the virus and host species are thought to be of importance. PBMCs were used in vitro to study the innate immune responses in sheep and goats in response to BTV infection [18]. Recently, global transcriptome analysis was used to understand the molecular events of host virus interactome in PPRV [19], Measles virus [20], [21] and Rinderpest virus [21] infections. However, to the best of our knowledge there are currently no reports on global transcriptome analysis of BTV-host interactome till date. Therefore, in the present study, as BTV is lymphotropic we infected the peripheral blood mononuclear cells (PBMCs) isolated from sheep and goats with BTV-16 virus and investigated the crosstalk vis-à-vis changes in global gene expression signatures at cellular level.
2. Results
2.1. Virus infection and confirmation
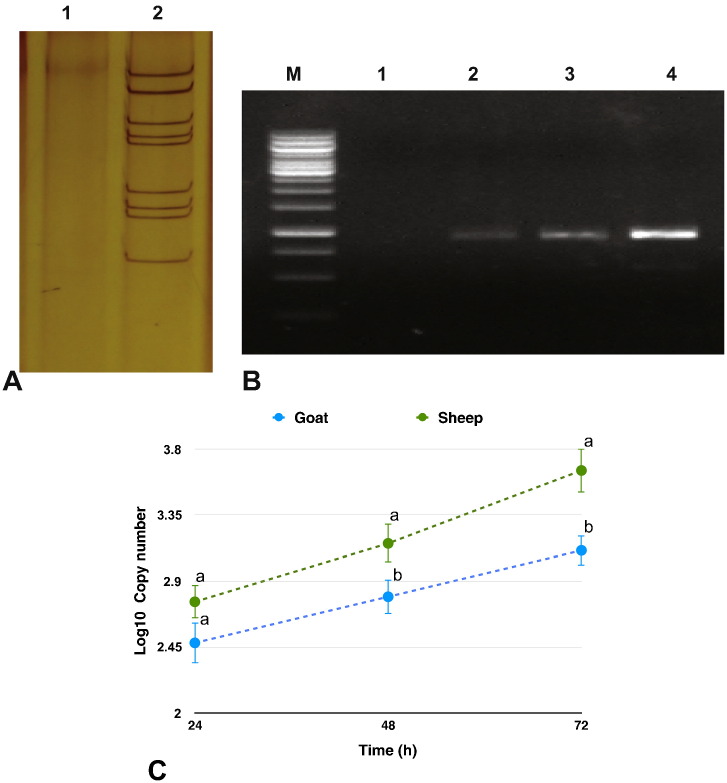
Virus grown in BHK-21 was confirmed for BTV infection. The purified virus at 105.0 TCID50/ml was used for infecting the sheep and goats PBMCs, independently. Viral infection was confirmed on amplification of 933 bp nonstructural 1 (NS1) gene fragment, at all time points - 24 h, 48 h, 72 h post infection (p.i.) (Fig. 1 A & B) in sheep and goats PBMCs. The cDNA from the mock-infected cells showed no amplification of the NS1 gene. Viral copy number across time points showed a gradual increase, being significantly highest at 72 h p.i., in both the species. At 48 h and 72 h p.i., the viral copy number was found to be significantly higher in sheep than in goats (Fig. 1C).
Fig. 1.
Confirmation of viral infection and viral transcript quantification: A) Amplification of NS1 gene by PCR at 24 h, 48 h and 72 h p.i in BTV-16 infected sheep PBMCs (M-1 kb DNA ladder; 1-Negative control; 2-Infected PBMC at 24 h p.i.; 3-Infected PBMC at 48 h p.i.; 4-Infected PBMC at 72 h p.i) B) Amplification of NS1 gene by PCR at 24 h, 48 h and 72 h p.i in BTV-16 infected goats PBMCs (M-1 kb DNA ladder; 1-Negative control; 2-Infected PBMC at 24 h p.i.; 3-Infected PBMC at 48 h p.i.; 4-Infected PBMC at 72 h p.i) C) Viral copy number across time points in sheep and goats by Real time PCR. Students t-test and Tukey HSD, were done in JMP 9.0., to compare the NS1 gene expression between sheep and goats at each time point and to compare the NS1 gene expression between time points within species, respectively. Levels not connected by same letter are significantly (P < 0.05) different within a time point between species. Across time points the viral copy number was significantly different between all time points in sheep. In goats, the viral copy number was found to be significantly different at 72 h p.i. w.r.t 24 and 48 h p.i.
2.2. Differentially expressed genes (DEGs) and functional analysis
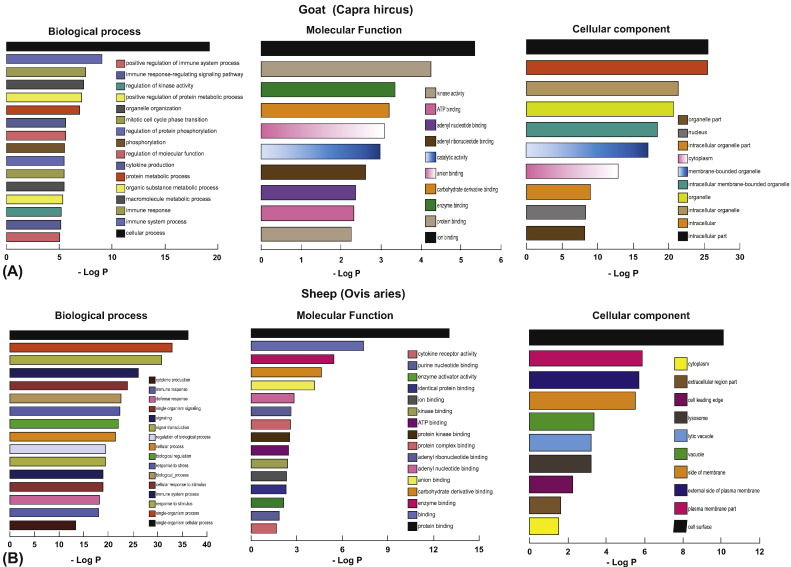
A total of 1152 and 2349 genes were differentially expressed in the infected PBMCs of sheep and goats, respectively, at 72 h p.i. Functional analysis of the differentially expressed genes was carried out using g:Profiler. GO terms categorized into Biological process (BP), Molecular function (MF) and Cellular component [22] were retrieved. In sheep, under the biological processes category, significant enrichment was seen for response to stimulus, immune system process, response to stress, cellular process, cytokine production, immune response, defense response, single organism signaling, etc. (Fig. 2A). Under molecular function, protein binding, enzyme binding, carbohydrate derivative binding, and anion binding were enriched in sheep (Fig. 2A). In the category of cellular component, increased numbers of DEGs were found enriched for cell surface, plasma membrane, external side of plasma membrane and lytic vacuole lysosome (Fig. 2A). Similarly, in goats, under biological processes, significant enrichment was seen for cellular process, immune system process, macromolecule metabolic process, organic substance metabolic process, cytokine production, regulation of molecular function, phosphorylation and organelle organization (Fig. 2B). Under the molecular function category, ion binding, protein binding, enzyme binding, carbohydrate derivative binding and anion binding were enriched in goats (Fig. 2B). In the category of cellular component, increased numbers of DE genes were enriched for intracellular part, intracellular organelle, intracellular membrane-bounded organelle and cytoplasm (Fig. 2B).
Fig. 2.
Functional enrichment of differentially expressed genes in BTV-16 infected sheep and goats PBMCs.
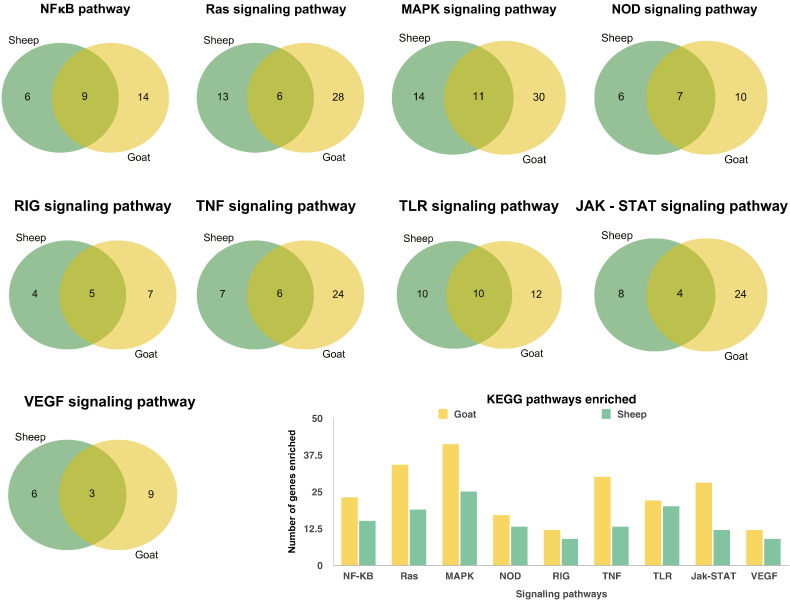
To further define differentially expressed gene function the biological pathways enriched in the infected transcriptomes were analyzed using KEGG database in ClueGo. Interestingly, the immune system processes were more enriched in goats than in sheep. In sheep and goats, Toll-like receptor, cytokine-cytokine receptor interaction/chemokine, NOD like receptor, Ras, MAPK, NOD like receptor, RIG, TNF, TLR, JAK-STAT, VEGF, B - cell and T- cell receptor, signaling pathways were found to be enriched on mapping of DEGs to canonical pathways in KEGG (Fig. 3). The MAPK signaling pathway was the pathway enriched with most number of genes both in sheep and goats. Among the DEGs enriched in immune processes, anti-viral genes mainly Interferon regulatory factors IRFs – 3 and 7; Interferon stimulated genes (ISGs) – IFIT1 (ISG56), IFIT2 (ISG54) and IFIT3 (ISG60); HERC5 and TLR7 were upregulated in sheep. TLRs- 4, 8 and 3; MAP2K and IKBKE were downregulated in sheep. In goats, IFITs − 1, 2 and 3; TLR7, HERC5, SMAD3, SMAD5, TGFBR1 and ITGA4 were upregulated. Some of the important immune genes are given in Table 1.
Fig. 3.
Pathways enriched in mapping the differentially expressed genes to canonical KEGG pathways in ClueGO. Venn diagrams show the genes involved in different pathways in sheep and goats. The bar chart indicates that more number of genes are involved in different pathways in goats than in sheep.
Table 1.
Fold change (Log2 fold change) of few immune genes in goat and sheep.
| Genes | Goat | Sheep |
|---|---|---|
| HERC5 | 1.20 | 1.94 |
| IFIT3 | 3.01 | 4.70 |
| IRF3 | – | 1.01 |
| STAT2 | 1.78 | 1.02 |
| IFIT1 | 3.71 | 4.28 |
| TLR7 | 2.33 | 1.41 |
| TLR4 | − 1.39 | − 3.53 |
| IRF7 | – | 1.75 |
| TLR8 | – | − 3.18 |
| TLR3 | – | 1.277 |
| IFIT2 | 4.3 | 3.16 |
| IFIT5 | 4.21 | 3.99 |
| ISG17 | 2.99 | 6.35 |
| ISG20 | – | 21.72 |
| SMAD3 | 1.94 | – |
| SMAD5 | 1.90 | – |
| TGFBR1 | 2.00 | – |
| ITGA4 | 2.39 | – |
2.3. Network analysis of differentially expressed genes in sheep and goats
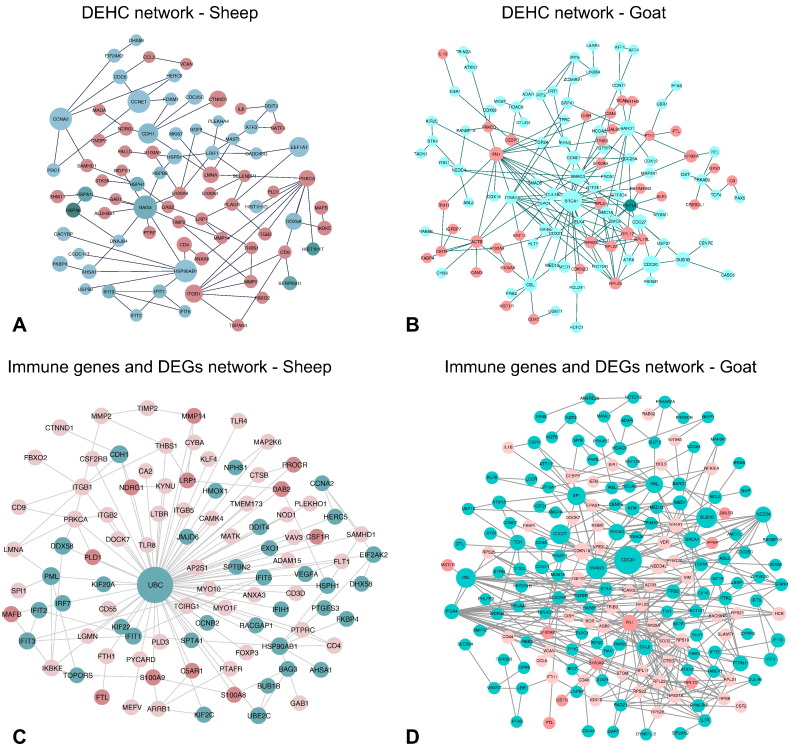
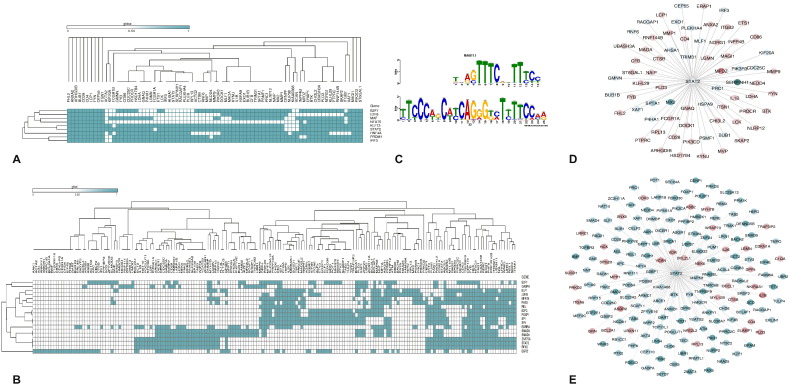
The protein-protein interaction network of the differentially expressed genes (DEGs) in sheep and goats was constructed from the available interactions in BioGRID database. The global protein interaction network of DEGs formed a dense hairball in both the species. The network involving only the DEGs was also found to be dense. To arrive at a representative network, the differentially expressed and highly connected (DEHC) genes (fold change > 3.5 and degree > 5) were considered. The DEHC network consisted of 192 genes in sheep and 207 genes in goats. Most of genes in the DEHC network were downregulated in sheep, but were upregulated in goats (Fig. 4 A & B). The DEGs involved in immune system processes in g:Profiler were further studied for their interactions with all the DEGs. A total of 308 and 470 DEGs were enriched in immune system process in sheep and goats, respectively. Out of these 308 and 470 immune process DEGs, 78 and 230; and 242 and 228 were up and down regulated, in sheep and goats, respectively. Interactome analysis of these immune process genes with all the DEGs resulted in 137 and 362 interactions involving a total of 109 and 180 DEGs, in sheep and goats. The network involving the interactions of the immune process genes and DEGs also shows most of the genes downregulated in sheep and upregulated in goats. Immune genes – TLR8, IKBKE, MAP2K, etc. were downregulated in sheep. However, ISGs viz. IFIT1, IFIT2 and IFIT3 responsible for anti-viral state in the host were upregulated in both sheep and goats.
Fig. 4.
Differentially expressed highly connected networks and immune - differentially expressed gene interactions in sheep and goats. The upregulated genes are shown in green and downregulated genes are shown in red with the gradient showing the extent of expression. The size of the node indicates connectivity (i.e. degree).
2.4. Transcription factors (TFs) co-regulating the expression of DEGs interacting with differentially expressed immune genes
The 5 kb upstream of 109 and 180 DE genes involved in immune processes in sheep and goats, was extracted to identify the common motifs conserved across these genes. These motifs were then submitted in the TOMTOM to identify TFs that co-regulate the expression of the DEGs. A total of 258 and 240 TFs were predicted to bind to the upstream of 109 and 180 DEGs in sheep and goats, respectively. Out of these TFs identified, 9 and 18 TFs were found to be differentially expressed in sheep and goats, respectively (Fig. 5 A & B). Out of the 18 TFs that were differentially expressed in goats, FOXP1, ELF1, GABPA, REL, SMAD3, STAT2, JUNB, ESRRA, SP1 and PAX5 were found to regulate immune system processes. TFs - JUNB and ESRRA were downregulated and the rest were upregulated. In sheep, IRF3 and STAT2 were the TFs enriched in immune system processes. Both these genes were found to be upregulated in sheep. Though IRF3 was not among the DEGs in goats it was found in the 240 TFs that co-regulate the immune DEGs. STAT2 was found to co-regulate 76 and 191 DEGs in sheep and goats, respectively (Fig. 5C, D & E). Most of the genes co-regulated by STAT2 were found to be downregulated in sheep but upregulated in goats.
Fig. 5.
Predicted transcription factors that co-regulate DEGs involved in immune processes: A) Heat Map showing the TFs binding to DEGs involved in immune processes in sheep. B) Heat Map showing the TFs binding to DEGs involved in immune processes in goats. The columns represent the DEGs involved in immune processes (Y-axis) and the rows represent the predicted TFs binding to the DEHC genes. C) Motif alignment of STAT2 that was commonly predicted to regulate DEGs involved in immune processes in sheep and goats D) network depicting the interaction of STAT2 with DE genes involved in immune processes in sheep E) Network depicting the interaction of STAT2 with DE genes involved in immune processes in goats.
2.5. Validation of RNA sequencing data by real-time RT-PCR
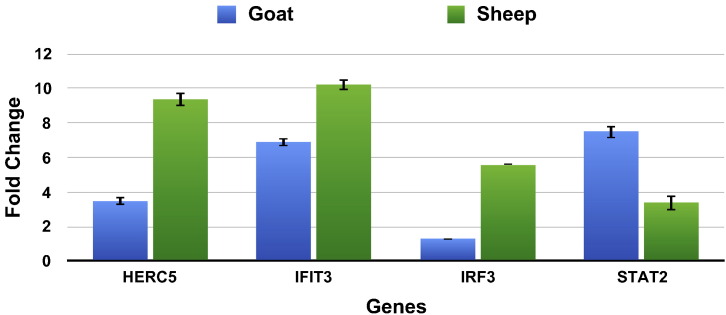
Four differentially expressed genes (HERC5, IRF3, IFIT3 and STAT2) involved in immune response pathways were validated for their expression using real time RT-PCR (Fig. 6, Table 2). The expression of these differentially expressed genes was in concordance with RNA sequencing results. IRF3 results corroborated with the RNA – Sequencing results in sheep. In goat, IRF3 was not in the DEG list after RNA – sequencing data analysis but was observed to have a fold change of 1.30 by real time PCR.
Fig. 6.
Quantitative real-time PCR to validate the RNA-seq experiment. Fold change (2− ΔΔCT) with control as the calibrator is represented along with the standard error of difference in both the species.
Table 2.
RNA – sequencing reads in sheep and goat samples.
| Parameter | Control goat R1 | Control goat R2 | Infected goat R1 | Infected goat R2 | Control sheep R1 | Control sheep R2 | Infected sheep R1 | Infected sheep R2 |
|---|---|---|---|---|---|---|---|---|
| Total no of reads | 30, 492, 162 | 30, 492, 162 | 30, 869, 892 | 30, 869, 892 | 38, 212, 792 | 38, 212, 792 | 23, 532, 326 | 23, 532, 326 |
| Read length | 101.00 | 101.00 | 101.00 | 101.00 | 101.00 | 101.00 | 101.00 | 101.00 |
| Total no. of good reads | 29, 112, 354 (95.47%) | 28, 886, 181 (94.73%) | 29, 494, 894 (95.55%) | 29, 212, 854 (94.63%) | 36, 072, 863 (94.40%) | 35, 745, 660 (93.54%) | 22, 202, 735 (94.35%) | 21, 934, 054 (93.21%) |
3. Discussion
BTV is an economically important pathogen of small ruminants. In India, the disease is more prevalent and severe in sheep, leading to high morbidity and the mortality rates in comparison to goats [23]. Antibodies to the virus (BTV) are common in cattle, buffaloes and goats, but the clinical disease has not been reported in these animals [24]. Recently, the RNA levels of the BTV in the PBMC culture supernatant showed, sheep PBMCs supports more replication of the virus compared to goats [18]. However, the severity of the disease in the sensitive hosts depends on the strain of the virus, breed, immune status of the animal, virulence of the virus, other host factors etc. Among the host factors, immune system plays an important role in response to the invading pathogens allowing the hosts to fight against these invaders and helping them to clear the pathogens from the system. Occasionally, pathogens manage to escape these barriers in certain hosts, establishing the infection more productively in comparison to other species. Differential innate immune responses elicited by different hosts in response to the virus infection play an important role in determining their susceptibility to the disease [25]. Till date, the molecular events underlying the differential disease causing potential of BTV in sheep and goats are poorly understood. High throughput approaches like RNA sequencing, helps in unravelling the candidate genes, pathways and the regulatory factors, contributing for differential species susceptibility. In this study the viral load was initially assessed at all time points (24 h, 48 h and 72 h p.i.) and was found to be significantly (P < 0.05) higher in both sheep and goats at 72 h p.i., and between species the expression was found significantly (P < 0.05) higher in sheep than in goats. Therefore, we investigated the global gene expression changes responsible for variable severity of the disease in sheep and goats at 72 h p.i. of BTV − 16 serotype.
Viruses target diverse signaling pathways in the host especially the immune signaling pathways, to evade host immune responses by altering the key molecules in the pathway through both physical and regulatory interactions [26]. BTV infection in sheep showed enrichment of pro-inflammatory pathways in pDCs of blood and anti-inflammatory pathways in pDCs of regional lymph nodes [27]. Our analysis revealed that BTV infection in PBMCs showed enrichment of RNA sensing RIG and other multiple host immune signaling pathways viz. NFκB signaling, MAPK signaling, Ras signaling, NOD signaling, RIG signaling, TNF signaling, TLR signaling, JAK-STAT signaling and VEGF signaling pathways. The core of BTV consists of the dsRNA genome and replication complexes encapsidated by VP3 dimer and VP7 trimer layers, which exhibit multiple solvent accessible translocation channels [28], VP3 and VP7 constitute the intermediate and inner capsid, respectively, and VP2 and VP5 constitute the outer capsid of BTV. On entry of the BTV virus into the cell, the outer capsid is removed, thus revealing the translocation channels leading to activation of transcription when necessary substrates are available in the cytosol. BTV cores produce ten positive sense ssRNA transcripts corresponding to each of the ten dsRNA segments [28], [29]. These transcripts are released from the core particle into the host cell cytoplasm where they act as templates both for translation and for negative strand viral RNA synthesis to generate genomic dsRNAs [28], [30], [31], [32]. The viral dsRNA is sensed by RIG-I and MDA5 in the cytosol, by TLR3 in the endosome compartment in innate immune cells, and by certain NLRs [33], [34]. The results in our study indicate that the presence of BTV genomic dsRNA in the cytosol must have triggered RIG and NOD like receptor signaling pathways, which further trigger NFκB and MAPK signaling pathways. MAPKs are key signaling molecules in innate immunity, which are widely conserved in eukaryotes and play an important role in response to many viral infections. The activation of MAPK signaling leads to the expression of cytokines or affects the growth of viruses [35]. Many RNA virus infections have shown differential expression of MAPK signaling pathway viz. enterovirus 71 [35], [36]. Also, the JAK/STAT pathway has also been shown to contribute to anti-viral immunity [37]. Among the immune signaling pathways enriched in our study, MAPK signaling pathway showed most significant enrichment in both the species under BTV infection. Involvement of more number of DEGs in MAPK and JAK-STAT signaling in goats indicates a possible better immune response in goats than in sheep.
The protein-protein interaction network of differentially expressed highly connected (DEHC) genes was dense in goats in comparison to sheep indicating greater degree of connectivity in goats among the DEHC genes. Further DEHC network analysis revealed majority of the DEHC genes were upregulated in goats when compared to the sheep. In the DEHC networks, IFITs (Interferon inducible tetricopeptides), the interferon stimulated genes responsible for establishing the anti-viral state [38] in the host were found to be upregulated both in sheep and goats. The relative higher expression of ISGs (IFIT1, IFIT5, MX1, OAS2 and ISG17) in sheep may be because of sheep PBMCs supporting higher virus replication than goats PBMCs as reported earlier [18]. TRIM family of proteins, that positively modulate innate immune signaling pathways triggered by pattern recognition receptors [39], [40], [41], [42]were found to be upregulated in goats. The network of differentially expressed immune genes with the other genes corroborated the above findings suggesting that the goats probably are better placed in responding to BTV in comparison to sheep.
Out of the transcription factors identified to co-regulate the immune DEGs, several of them were found to regulate immune system processes in goats. However, in sheep STAT2 and IRF3 were found to regulate the immune system processes. STAT2 was transcription factor identified commonly in sheep and goats. STAT2 plays a critical role in regulation of immune response [43]. The finding that most of the genes co-regulated by STAT2 were downregulated in sheep, but upregulated in goats reaffirms our previous finding. The dysregulation of genes in immune system processes with very few upregulated and many downregulated genes in sheep than in goats corresponded with significantly higher replication of BTV in sheep. The genes dysregulated and the networks perturbed in the present study indicate host variability with a positive shift in immune response to BTV in goats than in sheep.
4. Materials and methods
4.1. Ethics statement and animals
The experimental procedures in the present study were approved by Institute Animal Ethics Committee (I.A.E.C No.F.1.53/2012-13-J.D.). Goats and sheep (5 months old) used for blood collection were housed in appropriate containment facilities with feed and water ad libitum. Animals were screened for BTV antibodies using competitive ELISA (c-ELISA). PBMCs were isolated from blood collected from animals (n = 5) negative for BTV antibodies.
4.2. Virus and PBMC culture
BTV16 isolate grown in BHK-21 and purified (Ultracentifuged) was procured from National Typing Center laboratory, Department of Animal Biotechnology, LUVAS, Hisar. Geographically, the samples originated from Chennai (Tamil Nadu). Blood was collected from sheep and goats (n = 5 each) in heparin coated vacutainer vials. The whole blood was layered on 3 ml of histopaque-1077 and centrifuged at 2200 rpm for 40 min. The interphase rich in peripheral blood mononuclear cells (PBMCs) was transferred into separate tubes and washed three times with RPMI-1640 medium at 1800 rpm for 10 min. The final pellets were re-suspended in a complete medium containing RPMI-1640 and 10% fetal calf serum (FCS). Approximately, 2 × 106 cells per ml were seeded into two six well plates (for each species) for culture in 5% CO2 incubator at 37 °C.
4.3. PBMCs infection with BTV
PBMCs were seeded in two six well plates for each species separately. One plate served as a control (uninfected cells) and the other was used for infection. PBMC's were infected with BTV-16 at 105.0 TCID50/ml and incubated at 37 °C in 5% CO2 incubator for 72 h. Cells were observed for morphological changes like formation of viral inclusion bodies and virus specific tubules at 24 h, 48 h and 72 h p.i. Since non-structural 1 (NS1) gene is conserved in all BTV serotypes [10], PCR and Real time PCR for NS1 gene expression were done to confirm viral infection, at all time points. As the expression of NS1 was observed to be significantly higher at 72 h p.i. in both the species PBMCs at this time point were considered for RNA sequencing.
4.4. Library preparation and RNA sequencing
Infected cells were harvested at 72 h p.i. along with control (uninfected) cells. The harvested cells were pelleted at 2000 rpm for 10 min and stored in RNAlater at − 80 °C for RNA isolation. Total RNA was isolated from infected and mock-control cells using RNeasy Kit (Qiagen) according to the manufacturer's instructions. RNA was quantified and checked for RNA integrity number (RIN) using Agilent 2100 Bioanalyser. As the RIN number for both the infected and mock control was ≥ 7.0, mRNA was isolated using magnetic Oligo dT beads (Illumina) and purified using mRNA purification kit (Invitrogen). The purified mRNA from all the samples was then fragmented into small pieces (100–400 bp) using divalent cations for 5 min at 94 °C. The double stranded cDNA was first synthesized using Superscript double-stranded cDNA Synthesis Kit (Invitrogen, Camarillo, CA) using random hexamers (N6) primer (Illumina). The cDNA synthesized was then subjected to end repair and phosphorylation using T4 DNA polymerase, Klenow DNA polymerase and T4 phosphonucleotide Kinase. The end repaired cDNA fragments were then polyadenylated at the 3′end using Klenow Exo- 3′ - 5′ exo minus. Illumina paired end adapters were then ligated to the ends of the 3′ adenylated cDNA fragments. The adapters ligated cDNA was then enriched with PCR amplification using primer pairs (PE 1.0 and PE 2.0) (Illumina) catalyzed by Phusion DNA polymerase. The cDNA libraries prepared for both the control and the infected samples were sequenced on Illumina HiSeq 2000 platform according to the manufacturer's instructions. Illumina Sequencing was performed at Sandor Life Sciences Pvt. Ltd. (Hyderabad, India).
4.5. Transcriptome quantification and functional analysis of differentially expressed genes
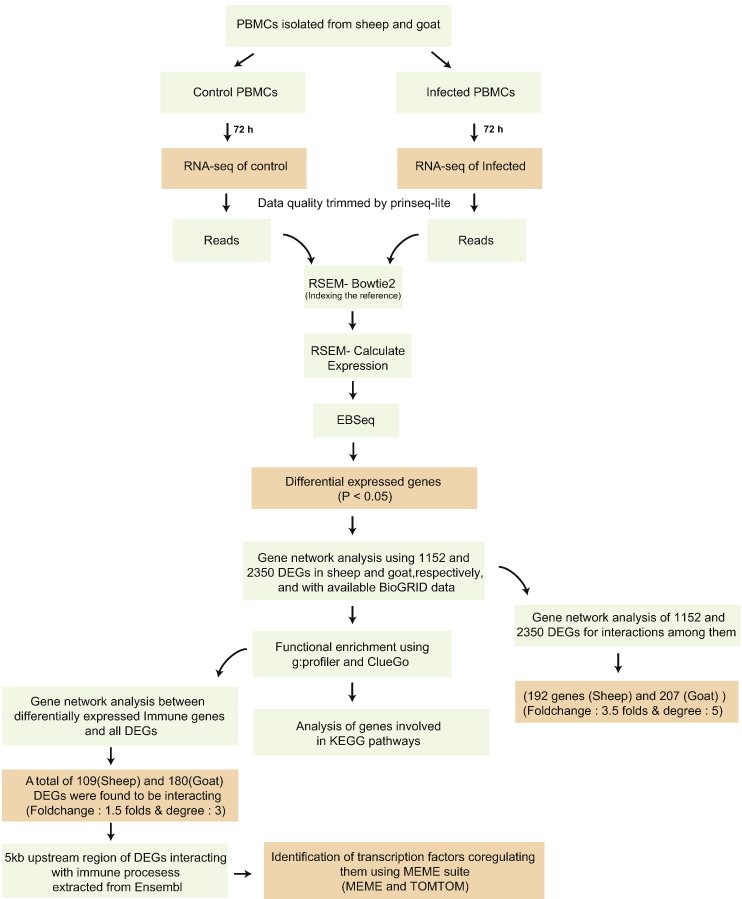
Fig. 7 highlights the various steps followed in the present study. The initial quality check of the sequencing data for control and infected samples was performed with NGS-QC toolkit. The number of reads per sample and their quality are given in Table 3.
Fig. 7.
Flow chart of steps depicting the analysis of transcriptome data.
Table 3.
List of primers used for Real time PCR in the study.
| HERC5 | Forward: GTATGAGGTTGGCTGGCATT | 198 bp |
|---|---|---|
| Reverse: CCCTGACTCCTCCAAAATCA | ||
| IRF3 | Forward: AGCGTCCCTAGCAGACAAGA | 220 bp |
| Reverse: CCAGGTTGAACACACCTCCT | ||
| IFIT3 | Forward: AAGGGTGGACACTGGTCAAG | 226 bp |
| Reverse: AGGGCCAGGAGAACTTTGAT | ||
| STAT2 | Forward: TGAATCACTGACTGCGGAAG | 155 bp |
| Reverse: CCAGAGTCAGGTAGCCGAAG | ||
| NS1 | Forward: CTTCTCTAGCTTGGCAACCACC | 274 bp |
| Reverse: AAGCCACCACTGTTTCCCGAT |
Reference genome of Ovis aries and corresponding gtf (gene transfer format) file were downloaded from Ensembl genome browser for further analysis. Transcript quantification was carried out with RSEM (RNA-Seq by expectation maximization), which assesses transcript abundances based on the mapping of RNA-Seq reads to the reference genome [44]. Briefly, maximum likelihood abundance estimates along with posterior mean estimates and 95% credibility intervals for genes/isoforms were calculated by RSEM. Two forms of abundance estimates are generated by RSEM, one, an estimate of the number of fragments that can be derived from an isoform or gene (the expected counts (EC)), and the other, the estimated fraction of transcripts within the sample that is represented by the given isoform or gene [45]. Differentially expressed genes (DEGs) in sheep and goats PBMCs infected with BTV, were identified using EBSeq package. The significantly (P ≤ 0.05) differentially expressed genes in sheep and goats, independently, were used for further analysis. Enrichment of Gene Ontology (GO) terms among the differentially expressed genes was analyzed using g:Profiler [46]. The DEGs were also mapped to KEGG database using ClueGo [47] to identify the biological pathways enriched among the DEGs.
4.6. Protein-protein interaction networks
The Biological General Repository for Interaction Datasets (BioGRID) is a curated biological database of protein-protein and genetic interactions created in 2003. It provides a comprehensive resource of protein–protein and genetic interactions for all major model organism species. BioGRID currently holds 557,158 non-redundant interactions and 804,431 raw interactions curated from both high-throughput data sets and individual focused studies derived from 44,686 publications in the primary literature (http://wiki.thebiogrid.org/doku.php/statistics). In this repository, protein-protein interactions in human are well defined and those in animal species are very few. Since protein interactions have been shown to be well-conserved across species [48]. g:Orth in g:Profiler web server was employed to produce orthology (functionally equivalent genes) predictions between species to facilitate functional and interaction annotation transfer across species [46]. To construct the protein-protein interaction network in the present study, Ovis aries orthologs in human were queried using g:Orth. Customized perl scripts were used to extract interactions involving the differentially expressed genes. The interaction network among the DEGs was visualized in Cytoscape 3.0.2 [49]. Degree or connectivity of a node/gene in a network is the number of connections it has to other nodes/genes. In sheep and goats 192 and 207 genes with fold change (positive or negative) > 3.0 and a degree of at least 5 were filtered from the DEGs to generate a sub-network using the original protein - protein interaction network, respectively. This sub network was named as differentially expressed and highly connected (DEHC) network. Gene network analysis between the differentially expressed immune genes with other DEGs was done considering a fold change of > 1.5 and degree of 3.0. A total of 109 and 108 DEGs were found to be involved in sheep and goats, respectively.
4.7. Extracting 5 kb upstream of immune DE genes
Ensembl is a comprehensive database addressing the challenges of decoding a eukaryotic genome from the set of functional elements it represents in providing access to the vast sea of data [22]. Ensemble BioMart is a hub for data retrieval across the taxonomic space. We extracted 5 kb upstream of 109 and 180 genes involved in immune processes in sheep and goats, respectively from Ensemble BioMart. The sequences containing a string of undefined bases (N′s) due to partially complete contigs were eliminated for further analysis using a customized perl script.
4.8. DNA motif discovery using MEME and prediction of transcription factors binding to upstream regions of immune DE genes
MEME (Multiple EM for Motif Elicitation) Suite is a comprehensive collection of tools most widely used for discovery of new transcription factors and protein domain binding sites [50]. The motif discovery algorithm MEME, finds ungapped motifs within DNA or protein sequences. TOMTOM [51] tool in the suite allows comparing the discovered motif to a database of motifs (JASPAR, JOLMA, Uni-PROBE, etc.) to find matches to the established Position Weight Matrices (PWMs) of transcription factors. To predict TFs that bind to the upstream of 109 and 180 genes identified in the present study, initially over-represented conserved motifs among these genes were identified using MEME followed by TOMTOM using a locally installed version of MEME suite. The TFs identified were mapped onto their orthologs in sheep. The presence of these TF binding sites across the immune process genes was depicted by a heatmap geneE software (Broad Institute).
4.9. Real – Time PCR
A total of four DEGs involved in immune response pathways were selected for further validation. Quantitative Real time PCR (qRT-PCR) was carried out on the same biological material that was used in RNA-seq experiment. RNA was extracted from the harvested cells using RNeasy mini kit (Qiagen) and was quantified using nanodrop spectrophotometer (Thermo Scientific). cDNA was synthesized using Revert Aid First Strand cDNA synthesis kit according to the manufacturer's instructions and qRT-PCR was performed using Applied Biosystems 7500 Fast system using 2 × SYBR Green Master mix (USB, Sigma). GAPDH was used as endogenous control for normalization of target gene(s) of interest. The primer sequences for the genes used for validation are given in Table 2. For all genes tenfold serial dilutions were run in the study to estimate the efficiency of PCR, and the percentage efficiency ranged between 95 and 100%. All the samples were run in triplicates. The relative expression of each sample was calculated using the 2− ΔΔCT method with control group as calibrator [52]. Viral copy number was estimated using NS1 gene of BTV (Primers in Table 1). A standard curve was initially constructed with ten fold serial dilutions and viral copy number was estimated using the equation - Ct = 37.326184–3.1831526 log10 copy no, where Ct is Mean NS1 cycle threshold at each time point. Students t-test and Tukey HSD, were done in JMP 9.0., to compare the NS1 gene expression between sheep and goats at each time point and to compare the NS1 gene expression between time points within species, respectively.
Statement of conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The funding for RNA sequencing was provided by Department of Biotechnology (BT/PR7729/AAQ/1/542/2013), Government of India and for qRT-PCR validation by Centre for Agricultural Bioinformatics (ICAR-IASRI).
Contributor Information
Anjali Singh, Email: anjalivet12@gmail.com.
Minakshi Prasad, Email: minakshi.abt@gmail.com.
Bina Mishra, Email: binachauhanmishra@hotmail.com.
Siddappa Manjunath, Email: smanju712@gmail.com.
Amit Ranjan Sahu, Email: dramitr.sahu@gmail.com.
G. Bhuvana Priya, Email: bhuvana.priya20@gmail.com.
Sajad Ahmad Wani, Email: wanisajad759@gmail.com.
Aditya Prasad Sahoo, Email: rush2aditya@gmail.com.
Amit Kumar, Email: vetamitchandan07@gmail.com.
Shweta Balodi, Email: shwetavet@gmail.com.
Anupama Deora, Email: shiningdeora@gmail.com.
Shikha Saxena, Email: saxena.shikha96@gmail.com.
Ravi Kumar Gandham, Email: gandham71@gmail.com.
References
- 1.Maclachlan N.J., Drew C.P., Darpel K.E., Worwa G. The pathology and pathogenesis of bluetongue. J. Comp. Pathol. 2009;141:1–16. doi: 10.1016/j.jcpa.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Osburn B.I., McGowan B., Heron B., Loomis E., Bushnell R., Stott J., Utterback W. Epizootiologic study of bluetongue: virologic and serologic results. Am. J. Vet. Res. 1981;42:884–887. [PubMed] [Google Scholar]
- 3.Howell P.G., Verwoerd D.W. Bluetongue virus. Virol. Monogr. Virusforschung Einzeldarstellungen. 1971;9:35–74. doi: 10.1007/978-3-7091-3987-5_2. [DOI] [PubMed] [Google Scholar]
- 4.Verwoerd D.W., Huismans H. Studies on the in vitro and the in vivo transcription of the bluetongue virus genome. Onderstepoort J. Vet. Res. 1972;39:185–191. [PubMed] [Google Scholar]
- 5.Verwoerd D.W., Louw H., Oellermann R.A. Characterization of bluetongue virus ribonucleic acid. J. Virol. 1970;5:1–7. doi: 10.1128/jvi.5.1.1-7.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratinier M., Caporale M., Golder M., Franzoni G., Allan K., Nunes S.F., Armezzani A., Bayoumy A., Rixon F., Shaw A., Palmarini M. Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog. 2011;7:e1002477. doi: 10.1371/journal.ppat.1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart M., Hardy A., Barry G., Pinto R.M., Caporale M., Melzi E., Hughes J., Taggart A., Janowicz A., Varela M., Ratinier M., Palmarini M. Characterization of a second open reading frame in genome segment 10 of bluetongue virus. J. Gen. Virol. 2015;96:3280–3293. doi: 10.1099/jgv.0.000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbs E.P., Greiner E.C. The epidemiology of bluetongue. Comp. Immunol. Microbiol. Infect. Dis. 1994;17:207–220. doi: 10.1016/0147-9571(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann M.A., Renzullo S., Mader M., Chaignat V., Worwa G., Thuer B. Genetic characterization of toggenburg orbivirus, a new bluetongue virus, from goats, Switzerland. Emerg. Infect. Dis. 2008;14:1855–1861. doi: 10.3201/eid1412.080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maan S., Maan N.S., Nomikou K., Veronesi E., Bachanek-Bankowska K., Belaganahalli M.N., Attoui H., Mertens P.P. Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS One. 2011;6:e26147. doi: 10.1371/journal.pone.0026147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenckel M., Breard E., Schulz C., Sailleau C., Viarouge C., Hoffmann B., Hoper D., Beer M., Zientara S. Complete coding genome sequence of putative novel bluetongue virus serotype 27. Genome Announc. 2015;3 doi: 10.1128/genomeA.00016-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niedbalski W. The evolution of bluetongue virus: genetic and phenotypic diversity of field strains. Pol. J. Vet. Sci. 2013;16:611–616. doi: 10.2478/pjvs-2013-0086. [DOI] [PubMed] [Google Scholar]
- 13.Pini A. Study on the pathogenesis of bluetongue: replication of the virus in the organs of infected sheep. Onderstepoort J. Vet. Res. 1976;43:159–164. [PubMed] [Google Scholar]
- 14.Darpel K.E., Monaghan P., Simpson J., Anthony S.J., Veronesi E., Brooks H.W., Elliott H., Brownlie J., Takamatsu H.H., Mellor P.S., Mertens P.P. Involvement of the skin during bluetongue virus infection and replication in the ruminant host. Vet. Res. 2012;43:40. doi: 10.1186/1297-9716-43-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barratt-Boyes S.M., Rossitto P.V., Stott J.L., MacLachlan N.J. Flow cytometric analysis of in vitro bluetongue virus infection of bovine blood mononuclear cells. J. Gen. Virol. 1992;73(Pt 8):1953–1960. doi: 10.1099/0022-1317-73-8-1953. [DOI] [PubMed] [Google Scholar]
- 16.Whetter L.E., Maclachlan N.J., Gebhard D.H., Heidner H.W., Moore P.F. Bluetongue virus infection of bovine monocytes. J. Gen. Virol. 1989;70(Pt 7):1663–1676. doi: 10.1099/0022-1317-70-7-1663. [DOI] [PubMed] [Google Scholar]
- 17.Stott J.L., Blanchard-Channell M., Scibienski R.J., Stott M.L. Interaction of bluetongue virus with bovine lymphocytes. J. Gen. Virol. 1990;71(Pt 2):363–368. doi: 10.1099/0022-1317-71-2-363. [DOI] [PubMed] [Google Scholar]
- 18.Dhanasekaran S., Vignesh A.R., Raj G.D., Reddy Y.K., Raja A., Tirumurugaan K.G. Comparative analysis of innate immune response following in vitro stimulation of sheep and goat peripheral blood mononuclear cells with bluetongue virus - serotype 23. Vet. Res. Commun. 2013;37:319–327. doi: 10.1007/s11259-013-9579-5. [DOI] [PubMed] [Google Scholar]
- 19.Manjunath S., Kumar G.R., Mishra B.P., Mishra B., Sahoo A.P., Joshi C.G., Tiwari A.K., Rajak K.K., Janga S.C. Genomic analysis of host - Peste des petits ruminants vaccine viral transcriptome uncovers transcription factors modulating immune regulatory pathways. Vet. Res. 2015;46:15. doi: 10.1186/s13567-015-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolt G., Berg K., Blixenkrone-Moller M. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 2002;83:1157–1165. doi: 10.1099/0022-1317-83-5-1157. [DOI] [PubMed] [Google Scholar]
- 21.Nanda S.K., Baron J., Royall E., Robinson L., Falciani F., Baron M.D. Infection of bovine dendritic cells by rinderpest or measles viruses induces different changes in host transcription. Virology. 2009;395:223–231. doi: 10.1016/j.virol.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Birney E., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Down T., Eyras E., Fernandez-Suarez X.M., Gane P., Gibbins B., Gilbert J., Hammond M., Hotz H.R., Iyer V., Jekosch K., Kahari A., Kasprzyk A., Keefe D., Keenan S., Lehvaslaiho H., McVicker G., Melsopp C., Meidl P., Mongin E., Pettett R., Potter S., Proctor G., Rae M., Searle S., Slater G., Smedley D., Smith J., Spooner W., Stabenau A., Stalker J., Storey R., Ureta-Vidal A., Woodwark K.C., Cameron G., Durbin R., Cox A., Hubbard T., Clamp M. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy P. Bluetongue virus: dissection of the polymerase complex. J. Gen. Virol. 2008;89:1789–1804. doi: 10.1099/vir.0.2008/002089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreenivasulu D., Subba Rao M.V., Reddy Y.N., Gard G.P. Overview of bluetongue disease, viruses, vectors, surveillance and unique features: the Indian sub-continent and adjacent regions. Vet. Ital. 2004;40:73–77. [PubMed] [Google Scholar]
- 25.Maclachlan N.J., Henderson C., Schwartz-Cornil I., Zientara S. The immune response of ruminant livestock to bluetongue virus: from type I interferon to antibody. Virus Res. 2014;182:71–77. doi: 10.1016/j.virusres.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 26.Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E., Hill D.E., Regev A., Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruscanu S., Jouneau L., Urien C., Bourge M., Lecardonnel J., Moroldo M., Loup B., Dalod M., Elhmouzi-Younes J., Bevilacqua C., Hope J., Vitour D., Zientara S., Meyer G., Schwartz-Cornil I. Dendritic cell subtypes from lymph nodes and blood show contrasted gene expression programs upon Bluetongue virus infection. J. Virol. 2013;87:9333–9343. doi: 10.1128/JVI.00631-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel A., Roy P. The molecular biology of Bluetongue virus replication. Virus Res. 2014;182:5–20. doi: 10.1016/j.virusres.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukusho A., Yu Y., Yamaguchi S., Roy P. Completion of the sequence of bluetongue virus serotype 10 by the characterization of a structural protein, VP6, and a non-structural protein, NS2. J. Gen. Virol. 1989;70(Pt 7):1677–1689. doi: 10.1099/0022-1317-70-7-1677. [DOI] [PubMed] [Google Scholar]
- 30.Mertens P.P., Brown F., Sangar D.V. Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology. 1984;135:207–217. doi: 10.1016/0042-6822(84)90131-4. [DOI] [PubMed] [Google Scholar]
- 31.Van Dijk A.A., Huismans H. The in vitro activation and further characterization of the bluetongue virus-associated transcriptase. Virology. 1980;104:347–356. doi: 10.1016/0042-6822(80)90339-6. [DOI] [PubMed] [Google Scholar]
- 32.Van Dijk A.A., Huismans H. In vitro transcription and translation of bluetongue virus mRNA. J. Gen. Virol. 1988;69(Pt 3):573–581. doi: 10.1099/0022-1317-69-3-573. [DOI] [PubMed] [Google Scholar]
- 33.Chauveau E., Doceul V., Lara E., Adam M., Breard E., Sailleau C., Viarouge C., Desprat A., Meyer G., Schwartz-Cornil I., Ruscanu S., Charley B., Zientara S., Vitour D. Sensing and control of bluetongue virus infection in epithelial cells via RIG-I and MDA5 helicases. J. Virol. 2012;86:11789–11799. doi: 10.1128/JVI.00430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen S., Thomsen A.R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi W., Hou X., Li X., Peng H., Shi M., Jiang Q., Liu X., Ji Y., Yao Y., He C., Lei X. Differential gene expressions of the MAPK signaling pathway in enterovirus 71-infected rhabdomyosarcoma cells. Braz. J. Infect. Dis. 2013;17:410–417. doi: 10.1016/j.bjid.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pleschka S., Wolff T., Ehrhardt C., Hobom G., Planz O., Rapp U.R., Ludwig S. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 2001;3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 37.Rauch I., Muller M., Decker T. The regulation of inflammation by interferons and their STATs. Jak-Stat. 2013;2:e23820. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Gack M.U., Kirchhofer A., Shin Y.C., Inn K.S., Liang C., Cui S., Myong S., Ha T., Hopfner K.P., Jung J.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNab F.W., Rajsbaum R., Stoye J.P., O'Garra A. Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 42.Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Au-Yeung N., Mandhana R., Horvath C.M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. Jak-Stat. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakasugi K., Crowhurst R.N., Bally J., Wood C.C., Hellens R.P., Waterhouse P.M. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One. 2013;8:e59534. doi: 10.1371/journal.pone.0059534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reimand J., Arak T., Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pages F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu H., Luscombe N.M., Lu H.X., Zhu X., Xia Y., Han J.D., Bertin N., Chung S., Vidal M., Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]