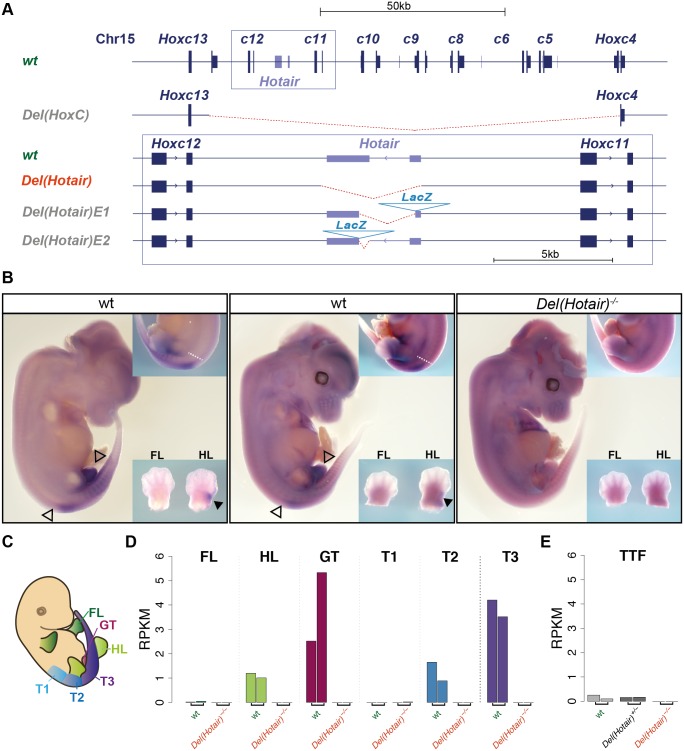
Fig 1. Hotair expression in vivo.
(A) Schematic representation of the wild type Hotair locus and the various Hotair deletion alleles. The deleted DNA is in red. The HoxC allele is from [18] and the shorter deletions in the box from [20] and [21]. (B) Whole mount in situ hybridization (WISH) of Hotair RNAs on E12.5 wild type CD1 (left) and CBA/C57/B6 (center) control embryos and of a Del(Hotair)-/- mouse embryo (right, n = 3). No signal was detected in Del(Hotair)-/- embryos, demonstrating the specificity of the probe. Hotair is expressed with a posterior restriction (white dashed line), resembling the transcript distribution of either a Hox11 or a Hox12 gene. Black arrowheads indicate expression domain of Hotair in the hindlimbs, hollow arrowheads indicate the limit of Hotair expression in the trunk and in the genital tubercle. The common artifact signal in the cerebral vesicles results from incomplete opening of these vesicles and subsequent probe trapping. (C) Schematic representation of the dissection patterns for RNA-seq. These dissections involved forelimbs (FL, dark green), hindlimbs (HL, green) and the genital tubercle (GT, magenta), as well as three trunk sections corresponding to the lumbar/sacral (T1, light blue); sacro/caudal (T2, blue) and caudal (T3, purple) regions. (D) Quantification of Hotair expression by RNA-seq (normalized RPKM values). (E) Quantification of Hotair expression (normalized RPKM values) in tail tip fibroblasts (TTF), using data from [20].