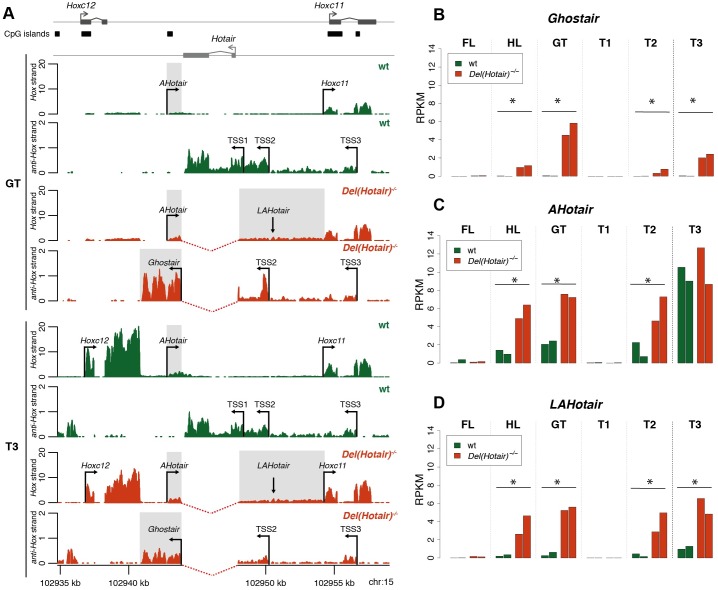
Fig 8. In-cis effects of the Hotair deletion on the local transcriptional activity.
A) RNA-seq expression profiles of the genomic region neighboring Hotair in both the developing genitalia (GT, four profiles on top) and the most posterior trunk tissue sample (T3, four profiles at the bottom) from either wild type (green) and Del(Hotair)-/- (orange) E12.5 embryos. In the wild type GT, only the Hoxc11 gene is expressed along with Hotair on the opposite strand, which shows at least three putative start sites (arrows, TSS1 to TSS3). In the mutant GT, a long form of a new lncRNA (AntiHotair) now extends (grey box) on the Hox DNA strand, going over the deleted region up to the Hoxc11 promoter. On the opposite DNA strand, the Hotair TSS2 and TSS3 are still functional and produce Ghost of Hotair (Ghostair), yet another new species of lncRNA, specific for the Del(Hotair)-/- allele (in orange) and absent from the control allele (in green). A similar situation is observed in the T3 trunk sample, except that Hoxc12 and AHotair are also expressed there. In the native locus, anti-Hotair is produced and meets with the end of the Hotair transcript. In the deleted allele, Ghostair is produced by the remaining Hotair TSS and terminates close to the 3’ end of the Hoxc12 transcript (bottom two profiles). The gray boxes indicate the genomic regions used for the expression quantifications of AHotair, LAHotair and Ghostair. The Y-axis represents the per-base RNA-seq read coverage, normalized by dividing by the total number of million mapped reads in the corresponding samples. The two biological replicates were pooled for this representation and only uniquely mapping reads were used. B-D) Expression values (normalized RPKM) for AHotair (B), LAHotair (C) and Ghost of Hotair (D) in all tissue samples. Genotypes are color-coded with wild type in green and Del(Hotair)-/- in orange. The asterisk* indicates those samples where significant differences in expression were scored between the two genotypes (FDR < 10%).