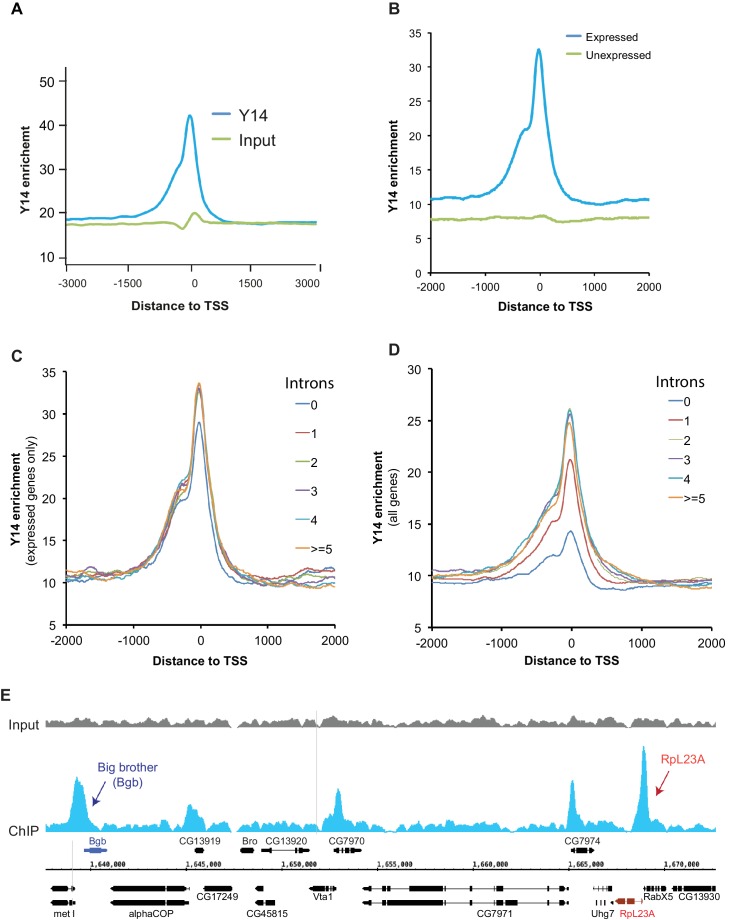
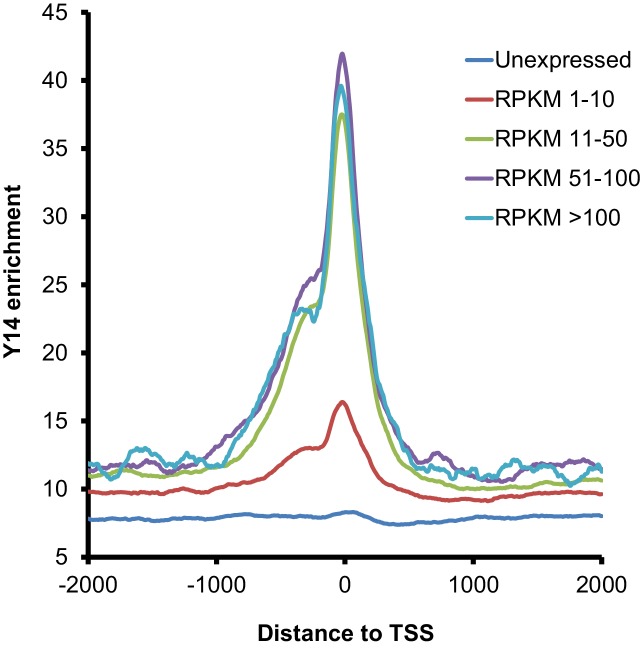
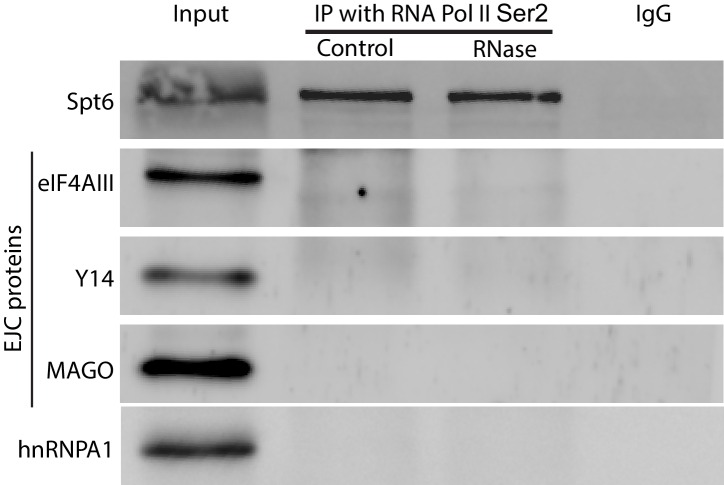
Figure 4. Y14 associates with expressed genes independent of introns in S2 cells.
(A) Genome-wide average Y14 ChIP-seq enrichment 3000 bases around transcription start sites (TSS) of ChIP (blue trace) versus input DNA (green). (B) Y14 ChIP-seq enrichment (after background subtraction) 2000 bases around transcription start sites (TSS) of expressed (blue trace) or unexpressed (green trace) genes in S2 cells (see Material and methods). (C) Average Y14 enrichment of expressed genes with a different number of introns: 0 to >=5, indicated by traces of different colour (see legend on right of the plot). (D) Average Y14 enrichment across genes as in C but including both expressed and unexpressed genes. (E) Representative chromosome region showing Y14 ChIP-seq enrichment profile (blue), versus that of input DNA (grey). Genes are labeled following Flybase nomenclature.