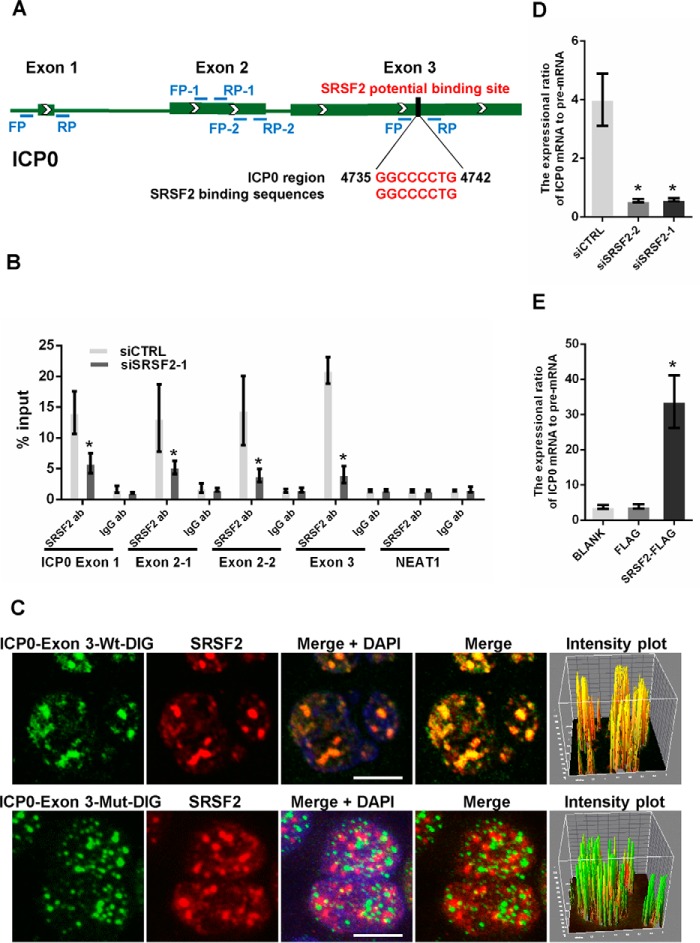
FIGURE 5.
SRSF2 regulated ICP0 pre-mRNA splicing. A, schematic of SRSF2 potential binding sites in the ICP0 pre-mRNA sequence. The black box shows the potential binding site, and red characters indicate matching sequences. B, after 36 h of transfection with the SRSF2 siRNAs or negative control, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. Cell lysates were harvested and subjected to an RNA immunoprecipitation assay. QRT-PCR was performed to detect the retrieval of ICP0 exon 1, ICP0 exon 2, ICP0 exon 3, and NEAT1 by the anti-SRSF2 antibody or anti-IgG antibody over the input level. The unrelated RNA NEAT1 was used as a negative control. The data are represented as the mean ± S.D. (*, p < 0.01, Student's t test). C, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h, fixed, incubated with DIG-labeled ICP0 exon 3 fragment (green) or ICP0 exon 3 fragment with a deletion in SRSF2 binding motifs (green) and SRSF2 (red) and subjected to confocal microscopy analysis. The intensity plots for the red and green channels were analyzed with the Image J software. DAPI (blue) was used to stain the nuclei. Scale bars, 10 μm. D, after 36 h of transfection with the SRSF2 siRNAs or negative control, the HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The expression ratio of the ICP0 mRNA to the pre-mRNA was determined by qRT-PCR in three independent experiments. The data are represented as the mean ± S.D. (*, p < 0.01, Student's t test). E, after 36 h of transfection with SRSF2-FLAG, FLAG, or BLANK, the HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The expression ratio of the ICP0 mRNA to pre-mRNA was determined by qRT-PCR in three independent experiments. The data are represented as the mean ± S.D. (*, p < 0.01, Student's t test).