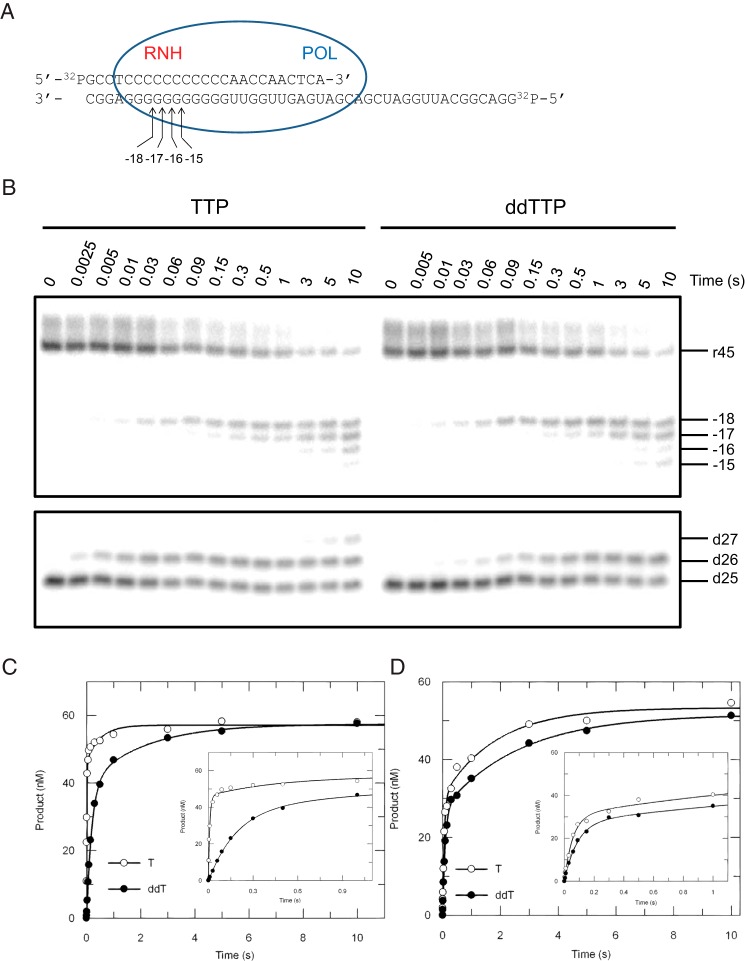
FIGURE 1.
Kinetics of nucleotide incorporation and RNase H cleavage. A, the “static” nomenclature for naming the cleavage sites are shown in the schematic; numbers refer to the distance from the primer terminus before polymerization. The sequence of the double-labeled DNA/RNA hybrid is shown. The blue oval represents HIVRT, where POL and RNH denote the polymerase and RNase H active sites, respectively. Positions of cleavage sites are counting from the 3′-terminus of the DNA primer, e.g. −18 means the cleavage site is 18 nucleotides upstream of the 3′-end of the DNA primer at the start of the reaction. B, to explore the potential coordination between the two active sites, polymerase (lower panel) and RNase H (upper panel) activities were measured simultaneously by preincubating 175 nm HIVRT with 75 nm double-labeled DNA/RNA hybrid and then mixing with 100 μm TTP or ddTTP in the presence of 10 mm Mg2+. Reactions were stopped by adding 0.5 m EDTA at various time points up to 10 s. The 45-nt RNA template and RNase H cleavages at positions −18 to −15 are marked as are positions of the 25-nt primer and extended products. Incorporation (C) and cleavage (D) data were analyzed by conventional fitting. The insets better display data points within 1 s. Nucleotide incorporation was fit to the double-exponential equation to derive rates of TTP or ddTTP incorporation as 107 ± 11 s−1 and 5 ± 0.3 s−1 for the fast phase and 2 ± 0.8 s−1 and 0.5 ± 0.1 s−1 for the slower phase, respectively. Similarly, rates of RNA cleavage during TTP or ddTTP incorporation were determined to be 17 ± 2 s−1 and 12 ± 0.7 s−1 for the fast phase and 0.5 ± 0.1 s−1 and 0.4 ± 0.04 s−1 for the slower phase.