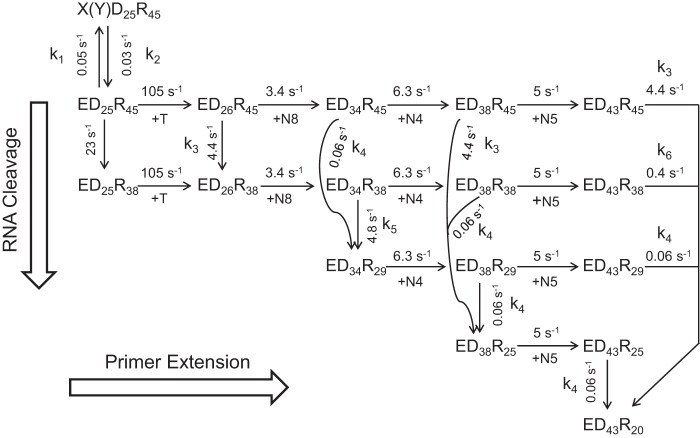
SCHEME 4.
Minimal model for processive polymerization and simultaneous RNase H cleavage. A minimal, abbreviated model describing processive polymerization and simultaneous RNase H cleavage is shown starting with the ED25R45 complex, where the subscript defines the length of each oligonucleotide. We also include the nonproductive complex: X(Y)D25R45 to quantitatively account for the amplitudes of the reactions. In this diagram polymerization steps go from left to right, whereas RNase H cleavage events go from top to bottom. Rate constants are given adjacent to the arrows for each reaction. The reaction starts with kinetic partitioning between polymerization to form D26 and RNase H to form R38; to describe the two activities as independent, we explicitly allow for either order of the two reactions. The next 8 nucleotide incorporations (+N8) are modeled as 1 step kinetically at 3.4 s−1 (8 nucleotides at 27 s−1 each). Subsequently, either the ED34R45 or the ED34R38 can be cleaved to form ED34R29 with the rate constants shown. Alternatively, either intermediate can be extended by 4 polymerization events (+N4) at a net rate of 6.3 s−1 (4 nucleotides at 25 s−1 each). Again, each elongation product can then either be extended by +N5 (at 5 s−1) or the RNA can be cleaved to form R25 (with different DNA states). Note that identical polymerization reactions are given identical rate constants independent of the state of the RNA, and identical RNase H cleavage events are given identical rate constants independent of the state of the DNA. This is an essential element of our model showing that the two reactions are independent. To simplify the model, we described several polymerization steps as occurring kinetically as a single step. Modeling the individual steps of polymerization gave the same results but required a much more complex model and computation.