Abstract
The MYC proto-oncogene is upregulated, often at the transcriptional level, in ∼80% of all cancers. MYC's promoter is governed by a higher order G-quadruplex (G4) structure in the NHE III1 region. Under a variety of conditions, multiple isoforms have been described to form from the first four continuous guanine runs (G41–4) predominating under the physiologically relevant supercoiled conditions. In the current study, short oligonucleotides complementing the 5′- and 3′-regions flanking the G4 have been connected by an abasic linker to form G4 clamps, varying both linker length and G4 isoform being targeted. Clamp A with an 18 Å linker was found to have marked affinity for its target isomer (G41–4) over the other major structures (G42–5 and G41–5, recognized by clamps B and C, respectively), and to be able to shift equilibrating DNA to foster greater G4 formation. In addition, clamp A, but not B or C, is able to modulate MYC promoter activity with a significant and dose-dependent effect on transcription driven by the Del4 plasmid. This linked clamp-mediated approach to G4 recognition represents a novel therapeutic mechanism with specificity for an individual promoter structure, amenable to a large array of promoters.
INTRODUCTION
MYC, a basic helix-loop-helix/leucine zipper transcription factor, is an enigmatic protein with over 30,000 potential binding sites in the human genome, 10–15% of which are generally bound at any one time. MYC has been shown to affect cellular proliferation, apoptosis, metastasis, angiogenesis and microenvironments (1). Disregulated MYC has been noted in vasculogenesis (2), restenosis (3,4), genomic instability (5–8) and proteolysis (9–12), but it is first and foremost known for its oncogenic role (13–16). MYC was one of the first proto-oncogenes to be described and is ultimately disregulated in most tumor types and stages.
MYC is a well-validated anti-cancer therapeutic target with no clinical compounds yet developed (17). The most notable progress in drug development related to MYC are bromodomain and extra terminal (BET) inhibitors, many of which are in clinical trials currently for hematological malignancies and some solid tumors (18–20). These compounds modulate MYC at the transcriptional level, which has been shown to be lethal to a variety of cancer types with a large potential therapeutic window and no long-term adverse effects on normal cells (21–24). Other means of effecting MYC mRNA expression include targeting the far upstream element (FUSE) (25–27) or non-canonical DNA structures in the proximal promoter, such as the G-quadruplex (G4) (28,29).
G4s are formed from G-rich regions of DNA, which are found to preferentially cluster around transcriptional start sites (TSS) throughout the genome. Such regions peak within 50 bp of the TSS (30,31), with a high prevalence in oncogenic promoters (32,33) including some representatives of the hallmarks of cancer (28). Negative superhelicity induced by transcription can promote local unwinding of these G-rich regions of DNA, which allows for the formation of G4s. G4s are made up of two or more stacked tetrads, formed by the Hoogsteen hydrogen bonding of four guanines, and are stabilized by monovalent cations, such as K+. Putative G4-forming regions have at least four runs of two or more consecutive guanines (G-tracts), most often three or more, separated by varying number of constitution of nucleotides that comprise the loop structures. The structures are classified by their loop directionality, length, and constitution. Formation of G4s in DNA has been shown to clearly form in vivo, where it modulates transcription (34–36); formation in RNA modulates translation and pre-mRNA splicing events (37–40). The unique globular structures of G4s, along with their potential to regulate the transcription of a host of oncogenes, make G4s an attractive drug target for the development of anti-cancer agents (30).
The most well-described oncogenic promoter containing a biologically active G4 is that within the MYC gene. Its near upstream core promoter region—the nuclease hypersensitivity element (NHE) III1—responsible for the initiation of 80–90% of transcription, adopts a predominantly parallel G4 that serves as a silencer element, putatively through sequestration of transcriptional factor binding sites (28,29). As one of the first described oncogenic promoter G4s in such an important oncogene, marked efforts have been made to identify a compound capable of stabilizing the MYC G4. Compounds have ranged from telomeric G4-stabilizers, natural products and DNA dyes, FDA approved drugs, metal complexes and aromatic fused heterocycles. These compounds have displayed in vivo anticancer activity in leukemia, lymphoma, medulloblastoma and cervical and nasopharyngeal cancers (41). Notably, through iterative fluorescence-based screenings, the ellipticine derivative NSC 338258 was identified as a stabilizer of the MYC G4 with nanomolar affinity (KD) and moderate selectivity, with in vitro activity demonstrated to be mediated by the promoter G4. However, NSC 338258 also demonstrated affinity for, and stabilization of, other similar promoter G4s, including those in the VEGF and HIF-1α genes (42). This work demonstrates the great potential for MYC G4-targeted therapies, but also the limitations with small molecule specificity.
To address the issue of specificity, nucleic acid (NA)-based approaches have been taken. In particular, the G4-forming, guanine-rich region, of the MYC promoter has been directly applied to leukemia cells with activity localized to both telomeric and non-telomeric DNA regions (43,44). NA's complementing the G- or C-rich region have been applied to cells to prevent duplex DNA formation, also mediating a decrease in promoter activity (45–47). Sequences complementing the G-rich strand and facilitating G4 formation, ex vivo, have led to DNA scission in a specific manner as well (48). In the works described herein, we took an NA-based approach where we complemented the DNA region on the 5′ and 3′-flanks of the G4-forming region and restricted the distance between the two flank complements to ∼18 Å. This approach enabled us to maintain a particular G4 isoform within the MYC NHE III1 region, and to be very specific to only the MYC promoter in a reversible manner. The developed NA-clamps demonstrate recognition of the MYC G4 and are able to facilitate the formation of the higher order structure, leading to a downregulation of promoter activity.
MATERIALS AND METHODS
Materials
Oligonucleotides and nucleic acid (NA) clamps (Table 1) were purchased from Eurofins MWG Operon (Huntsville, AL, USA). Unless otherwise noted, all other chemicals were purchased from either Thermo Fisher Scientific (Waltham, MA, USA) or Sigma-Aldrich (St. Louis, MO, USA).
Table 1. Oligonucleotides used in the current study.
| Name | Sequence (5′-3′) |
|---|---|
| Pu46 | [6FAM]GCGCTTATGGGGAGGGTGGGGAGGGTGGGGAAGGTGGGGAGGAGAC |
| Pu46 (unlabeled) | GCGCTTATGGGGAGGGTGGGGAGGGTGGGGAAGGTGGGGAGGAGAC |
| Py46 | GTCTCCTCCCCACCTTCCCCACCCTCCCCACCCTCCCCATAAGCGC |
| mt 5 | [6FAM]GCGCTTATGGGGAGGGTGGGGAGGGTGTGGAAGGTGGGGAGGAGAC |
| mt 5 (unlabeled) | GCGCTTATGGGGAGGGTGGGGAGGGTGTGGAAGGTGGGGAGGAGAC |
| mt 1,6 | [6FAM]GCGCTTATGGTGAGGGTGGGGAGGGTGGGGAAGGTGTGGAGGAGAC |
| mt 1,6 (unlabeled) | GCGCTTATGGTGAGGGTGGGGAGGGTGGGGAAGGTGTGGAGGAGAC |
| mt3 | [6FAM]GCGCTTATGGGGAGGGTTTTTAGGGTGGGGAAGGTGGGGAGGAGAC |
| mt 1,2,3,4 | [6FAM]GCGCTTATGGTGAGTGTGGTGAGTGTGGGGAAGGTGGGGAGGAGAC |
| mt 1,2,3,4 (unlabeled) | GCGCTTATGGTGAGTGTGGTGAGTGTGGGGAAGGTGGGGAGGAGAC |
| Clamp A (A1) | CTCCTCCCCACCTTCCCC[C-18]ATAAG |
| Clamp A2 | CTCCTCCCCACCTTCCC[C-18][C-3]TAAG |
| Clamp A3 | CTCCTCCCCACCTTCC[C-18][C-3][C-9]AAG |
| Clamp B | CTGAGTCTCCTCCCCACCTT [C-18]TCCCC |
| Clamp C | CTGAGTCTCCTCCCCACCTT [C-18]ATAAG |
| 5′ | CTCCTCCCCACCTTCCCC |
| 3′ | ATAAG |
| 5′ link | CTCCTCCCCACCTTCCCC[C-18] |
| 3′ link | [C-18]ATAAG |
Electrophoretic mobility shift assay (EMSA)
Fluorescent oligonucleotides were formamide treated (100% for 15 min at 95°C; snap cooled on ice) and purified on a 10% polyacrylamide (from acrylamide/bisacrylamide solution (29:1)) gel. 5′-6-FAM labeled oligonucleotides (1 μM) were dissolved in Tris-HCl buffer (50 mM, pH 7.4) with KCl (25 mM) and annealed by heating to 95°C for 5 min before slowly cooling to 25°C. Linked nucleic acid clamps (varying concentration denoted in each experiment) were added either pre- or post-annealing; if they were added post-annealing, they were incubated for 30 min. As noted in the text, unlabeled oligonucleotides were added to this 30 min incubation to compete for clamp binding. dsDNA of the Pu46 sequence was formed by annealing the G-rich Pu46 sequence with its complementary C-rich Py46 sequence. Samples were loaded on a 10% non-denaturing polyacrylamide-TBE gel and electrophoresed at 150 V for 1 h. Banding was visualized using a FotoDyne Analyst Investigator FX documentation system equipped with Red/Green/Blue LED epifluoresence. Quantitation of banding was performed with ImageJ software (Bethesda, MD, USA) to determine the optical density (OD) of each band.
Electronic circular dichroism (ECD)
Non-FAM labeled oligonucleotides were dissolved in Tris–HCl buffer (50 mM, pH 7) to a final concentration of 5 μM with 25 mM KCl. The oligonucleotides were annealed as described above; ECD spectra were recorded on a Olis DSM-20 spectropolarimeter (Huntsville, AL, USA), using a quartz cell of 1 mm optical path length and a response time as a function of high volts, spanning wavelength 225–350 nm.
Transfection and luciferase
The human embryonic kidney (HEK)-293 cell line was obtained form ATCC (Manassas, VA, USA) and was cultured in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin solution. Cell culture were maintained at 37°C in a humid incubator supplied with 5% CO2. HEK-293 cells in exponential growth stage were used for transfection. Briefly, cells were seeded at 8 × 104 cells/well in 24-well plates and allowed to attach overnight before being used for transfection. Cells were then co-transfected with either the Del4 (250 ng) or the promoterless empty vector plasmid pGL4.17 (250 ng) plus renilla plasmid (pRL-SV40, 100 ng), either with or without NA clamps, using Fugene HD in a 3:1 ratio. 24 or 48 h after transfection, cells were lysed in passive lysis buffer (Promega), taken through two freeze/thaw cycles to promote cell lysis, and luciferase was measured with the Dual Luciferase Assay kit (Promega) using a Lumat LB9507 luminometer. Luciferase experiments were replicated three times with internal duplicates measured within each experiment. Fold-changes induced by the clamps in the Del4 plasmid were normalized to any effects noted in the EV plasmid, as in (49). One-way ANOVA with post-hoc Tukey's test was employed to determine the statistical significance. The pGL4.17 plasmid, Fugene HD, and dual luciferase assay kit were purchased from Promega (Madison, WI); the Del4 luciferase plasmid driven by MYC promoter was a gift from Burt Vogelstein (Addgene plasmid #16604, Cambridge, MA, USA)(50).
Dimethyl sulfate (DMS) footprinting
FAM-labeled Pu46 (10 μM) either with or without clamp (10 μM) was added to a buffer of 25 mM KCl and 50 mM of Tris–HCl. Dimethylsulphate (DMS, 10%) in 40% ethanol was added to each sample to a final concentration of 0.5% DMS and 2% ethanol. The solution was incubated for 15 min before adding calf-thymus DNA (1 μg) and gel loading buffer (bromophenol blue, 0.005% final concentration). Samples were then subjected to electrophoresis as described above with EMSA, and the isolated bands were excised and extracted with gel elution buffer (0.4 M ammonium acetate, 1 mM MgCl2, 0.2% SDS) in a 37°C water bath overnight. The supernatant was collected and DNA was precipitated in 75% ethanol with 3 M sodium acetate at -20°C overnight; samples were centrifuged at 15 000 rpm for 30 min and the supernatant was discarded, DNA pellets were air-dried for 30 min before they were re-suspended in 10% piperidine to cleave DMS tagged DNA, heated at 95°C for 30 min and snap-cooled on ice. DNA was precipitated in 75% ethanol/3M sodium acetate as above, then dried and washed twice with water. Finally, DNA was suspended in 15 μl of DMS dye (80% formamide, 10 mM NaOH, 0.005% bromophenol blue), heated to 85°C for 3 min, cooled on ice immediately and run on a 16% denaturing gel with 7 M urea at 2 W, overnight at 4°C. The gel was visualized as described above. Image J software (Bethesda, MD, USA) was used to develop histograms if the guanine protection pattern.
RESULTS
NA-clamp binding and selectivity
Clamp theory and linker length optimization
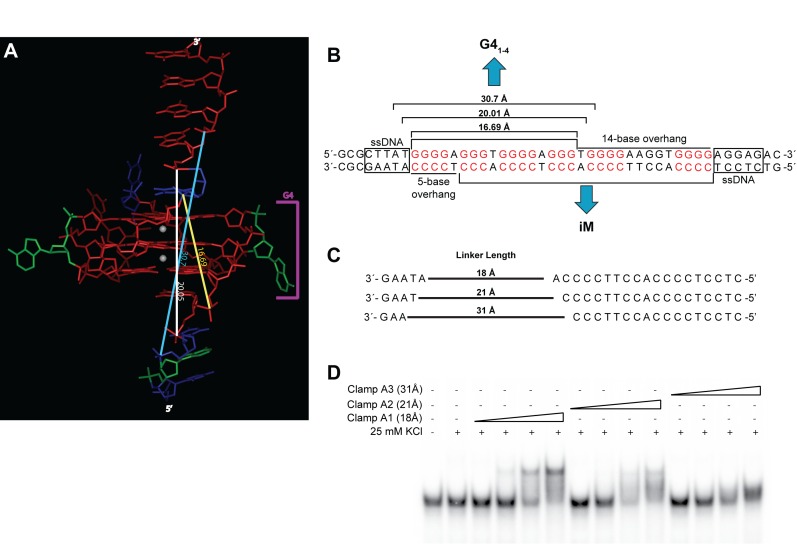
The predominant G4 isoform identified under supercoiling conditions (51), G41–4, was modeled with both 3′- and 5′-flanking regions in order to determine the 3D space occupied by the higher order DNA structure. Linearly, the 16 nucleotide (nt) sequence, with loops of 1, 2 and 1 nt, respectively in the 5′-3′ direction, occupies ∼58 Å. When the G4 is formed, the immediate proximal nt on the 5′- and 3′-side of the G4 measured 16.7 Å apart, the second most proximal nt measured 20.1 Å apart, and the third most proximal nt in each direction measured 30.7 Å apart (Figure 1A). Accordingly, complementary NA-clamps were designed to complement the appropriate flanking nt in the 5′- and 3′- directions with polyethylene glycol phosphate linkers connecting the regions of 17.7 Å (clamp A1), 21.3 Å (clamp A2) or 31 Å (clamp A3) (Figure 1B and C). Each clamp was examined for the ability to bind the wild-type (WT) Pu46 sequence by EMSA; although all clamps demonstrated a dose-dependent ability to retard the migration of the MYC G4 sequence, clamp A1 showed the greatest affinity for the sequence (Figure 1D), now referred to as ‘clamp A’; all future clamp linkers were maintained at 17.7 Å.
Figure 1.
Three-dimensional analysis of the MYC NHE III1 G41–4 structure and clamp-mediated recognition. (A) Computer modelling of the G4 structure identified in supercoiled DNA (46) examined the distance between the first (16.69 Å, yellow), second (20.05 Å, white) or third (30.7 Å, light blue) proximal bases. (B) G4 and iM offset in supercoiled DNA (46) mediates varying length unpaired G4 flanking regions that were used to (C) design clamps to recognize and bind the MYC G4. (D) EMSA analysis of clamp binding (0.01−10 μM) to the MYC WT (Pu46) sequence highlighted greater recognition with shorter linkers. The best binding occurs with the 5′- and 3′- complementary sequences linked by 18 Å.
Clamp isoform selectivity
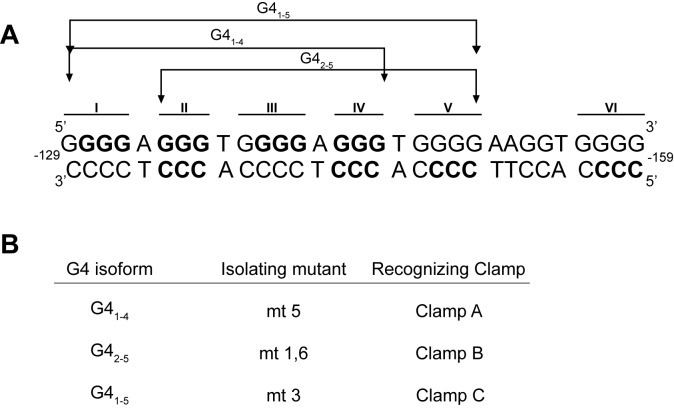
In addition to the G4 isoform noted under supercoiled conditions (G41–4), two other all parallel structures have been noted to form from linear DNA: G42–5, with loops of 1:2:1 in the 5′-3′ direction (52,53) and G41–5, with loops of 1:6:1 in the 5′-3′ direction (54). Keeping the linker distance locked at ∼18 Å, two additional clamps, B and C, were designed to recognize the additional two G4s, respectively. Additionally, the Pu46 sequence was mutated with G-to-T transitions in order isolate an individual predominant isoform from the WT sequence (Figure 2A and B).
Figure 2.
Clamp development for multiple G4 structures in the MYC promoter. (A) From the six runs of three or more continuous guanines, several structures have been described. Under supercoiled conditions, the nucleotides involved in the major G4 and iM structure are bolded 51. (B) A number of G-to-T mutant sequences were developed to isolate each structure, and clamps with 18 Å linkers were designed to recognize each major isoform.
Each of these clamps was incubated with the WT sequence, post-annealing and examined by EMSA (Figure 3A, left). In this and all future EMSAs, band clarity was enhanced and it is possible to visualize multiple upwardly shifted bands, particularly with clamp A, which recognizes G41–4. Only one slower migrating band is visible with clamps B and C. Clamp A, however, demonstrates a dose-dependent shift from no binding, to recognizing two conformations of the DNA, to only one band at 10 μM. Using a complete G4 knockout sequence (mt 1–4), binding of each clamp was also observed (Figure 3A, right). The slower migration pattern of clamps B and C did not change as compared to WT; only the lower migrating band was observed with clamp A. As this sequence is unable to form a G4 but maintains the complementary flanking regions, this lower migrating DNA band represents the clamp complementing the linear sequence and forming dsDNA, but not the higher order G4 structure.
Figure 3.
Binding and specificity of linked clamps to MYC G4s. (A) The binding of clamps A-C was examined in a dose-dependent manner to the WT and a G4 knockout mutant sequence (mt 1,2,3,4). Two upwardly shifted bands were noted with clamp A, but only one slower migrating band was visible with clamp B or C, with the WT sequence. The lower band with clamp A and the single band with clamps B and C were also noted in the mt 1,2,3,4 EMSA, suggesting that the clamps are still complementing the flanking regions, but are not recognizing a G4 structure. Only clamp A facilitates the supra-shifted DNA band putatively containing a G4 clamped by complementing the 5′- and 3′-flanking regions. (B) ECD confirms parallel G4 structures (spectral maxima ∼260-265 nm) are formed from the WT and mt 5, but not the mt 1,2,3,4, sequences (black lines). Upon the addition of equimolar clamp a (blue lines), G4 cotton effect are maintained and the spectra widen to encompass dsDNA regions of 275 nm with the WT and mt 5 sequences; mt 1,2,3,4 with clamp A demonstrates dsDNA cotton effects only and supports the moderate shifted band in (A) representing clamp binding to the flanking regions only. (C) Clamp A is designed to recognize G41–4, isolated with mt 5. The supra-shifted EMSA band is evident with WT and mt 5 only and the moderately shifted complementary band is noted with mt 1,6 (isolated G42–5) and mt 3 (isolating G41–6). This confirms the isoform specificity of clamp A. (D) Incubation of clamp A with WT MYC G4 before (pre-), versus after (post-) annealing fosters more supra-shifted banding (band i) in a dose dependent manner, most notably at 1 μM. This is compared with the moderately shifted dsDNA band (ii) and the linear/G4 band (iii). Semi-quantitation (below) of the bands with ImageJ reveals an ∼2.5-fold increase in G4 formation when clamp A (1 μM) is incubated pre-annealing.
dsDNA formation, or a mixture of dsDNA and G4 formation, of the WT, mt 5 or mt 1,2,3,4 sequences in the presence of clamp A was examined by ECD (Figure 3B). In the presence of equimolar clamp A, both the WT and mt 5 sequences maintained their signature parallel maxima at 265 nm (also noted in the absence of clamp), while also broadening to demonstrate a cotton effect around 275 nm, which is consistent with dsDNA. The mt 1,2,3,4 sequence shifted from a nondescript spectral signature in the absence of clamp to clear dsDNA maxima with a broad cotton effect at 275 nm. These ECD confirmed the moderately slower migrating band noted with clamps B and C and with all mt 1,2,3,4 EMSAs to represent clamp binding to the sequence and facilitating dsDNA but no G4. In contrast, the spectra noted with WT and mt 5 sequences in the presence of clamp A confirmed that the most upwardly migrated band contains a mixture of G4 and dsDNA wherein the clamps complement the flanking regions and restrain the G4 structure.
Clamp A was designed to be selective for the G41–4 structure, and not to recognize the G42–5 or G41–5 isoforms. Thus its binding was evaluated to multiple sequences: WT, which forms multiple structures in equilibrium, mt 5, which isolates G41–4, mt 1,6, which isolates G42–5, and mt 3, which isolates G41–5 (Figure 3C). Clamp A (10 μM) migrated to the highest supershift (termed supra-shift) with the WT and mt 5, but not the mt 1,6 or mt 3 sequences, confirming that it is only recognizing its target G4 isoform (G41–4). Complementary binding facilitating dsDNA mediating a moderate upward band shift is noted with the mt 1,6 and mt 3 sequences.
G41–4 recognition and induction
Incubating the clamps with the G-rich sequences after they are annealed into G4s is a measure of structure recognition. We additionally sought to study if the clamps can participate in ssDNA ←→ G4 equilibrium by annealing the G-rich sequences in the presence of the clamp (‘pre’) and comparing that with adding the clamp after G4s are formed (‘post’) (Figure 3D). Band density was quantitated with Image J software, and each band was expressed as a fraction of the total OD. Band i is the supra-shifted G4:dsDNA entity, band ii is the dsDNA structure, and the sum of lower bands combined in iii represent the linear and unbound G4 bands. Notably, 1 μM of clamp A incubated with the WT DNA pre-annealing (lane 3), as compared to post-annealing (lane 6), facilitated more G4-formation as evidenced by more supra-shifted banding (i). In lane 3, bands i, ii, and iii exist in ratios of 0.44, 0.44 and 0.12, respectively, whereas in lane 6 they are in ratios of 0.18, 0.55 and 0.26, respectively. Thus, G4 formation is increased approximately 2.5-fold.
Clamp A G4 affinity and structure facilitation
Preferential G41–4 affinity
Recognition and binding of clamp A to the particular isoform it was designed towards, namely G41–4, was examined in a series of competition EMSA's. While maintaining 1:1 binding of the WT MYC sequence and clamp A, various unlabelled competing oligonucleotides were added to the incubating samples, post-annealing (Figure 4). dsDNA, even at a 5-fold excess, was unable to compete off any clamp binding. Only at that 5-fold excess were the linear mt 1,2,3,4 and the alternative G4 isoform-forming mt 1,6 able to decrease G4 recognition of clamp A from the WT sequence, as noted with the disappearance of the supra-shifted band. In contrast, at an equimolar ratio of unlabelled mt 5, which is capable of only forming G41–4, the supra-shifted band is markedly decreased (Figure 4A). A more narrow range of competing mt 5 was examined in order to probe the general affinity of clamp A for its target structure from the mixture of equilibrating G4s forming with the WT sequence (Figure 4B). mt 5 was able to compete off clamp A from the WT G4 at ratios as low as 0.5, and completely by a ratio of 1.2. This highlights the high recognition of clamp A for its target sequence G41–4.
Figure 4.
Clamp A has highest affinity for G41–4. (A) Clamp A affinity for the WT MYC G4 was examined by competing for binding by various sequences/structures by EMSA. dsDNA of the NHE III1 region was unable to compete the clamp away from the WT G4 at even 5 fold-excess, whereas both the linear mt 1,2,3,4 and the alternate structure formed by mt 1,6 (G42–5) were able to decrease the supra-shifted banding at 5 μM. Most notably, mt 5, which isolates the target of clamp A (G41–4) is able to compete away the supra-shifted band at a 1:1 ratio. (B) More narrow ranges of competing mt 5 for binding clamp A away from WT MYC G4 reveals a decrease in the suprashifted band, and thus a preference for the single G4 species (G41–4) from the milieu of competing structures at <1 μM. (C) Partitioned fragments of Clamp A were compared to the intact clamp for post-annealing binding to Pu46 by EMSA. Notable shifted banding, and particularly the presence of the supra-shifted band, were evident only with the intact Clamp A.
Clamp composition
Clamp A is composed of three individual fragments—a five nucleotide portion complementing the 5′ end of Pu46 (the 3′ end of the clamp), an 18 nucleotide portion complementing the 3′ end of Pu46 (the 5′ end of the clamp), and a 17.7 Å polyethylene glycol phosphate linker connecting the two complementary ends. In order to examine what portion(s) of the clamp are necessary for G4 recognition, each individual fragment was examined for binding to Pu46 post-annealing (Figure 4C). The longer 5′ end (alone or with the linker region attached) is able to complement Pu46 better than the shorter 3′ end at an equivalent molarity. When both the 5′ and 3′ clamp fragments are present, either with or without any linkers attached, there is a higher shift than either portion alone. Comparing Clamp A binding (lane 2) to the 5′ link + 3′ (lane 8) or 5′ + 3′ link (lane 9) patterns, each of which contain all fragments necessary for higher order structure recognition, it is evident that supershifts only occur when all three fragments are present and physically connected.
Facilitating G41–4 formation
To confirm that the G4 isoform recognized by clamp A is G41–4, bands were extracted from an EMSA gel (Figure 5A) and were subjected to chemical (DMS) footprinting (Figure 5B). The G4 isolated from Pu46 not incubated with Clamp A, in DMS lane i, matching band i extracted from EMSA, is a mixture of species, with clear protection in runs 2−4, and less discriminate patterns of protection in runs 1 and 5. Band ii represents linear DNA as evidenced by no apparent guanine protection pattern evident post-DMS labelling. Band iii is a mixture of linear and G4 structures that are not G41–4, and an incomplete protection pattern is evident through multiple guanine runs. In lane iv, a pattern of guanine protection is noted in runs 1–4, particularly matching the intended structure that was previously noted in supercoiled DNA (51).
Figure 5.
G4 structural variation in unbound, versus clamp bound, MYC WT DNA. (A) Bands were separated by EMSA, extracted, DMS treated, and run on a sequencing gel (B). Pu46 + 25 mM KCl with no clamp A (band i) forms an array of equilibrating sequences with clear patterns of protection in runs 2 and 4, and partial patterns of protection in runs 1, 3 and 5. Upon the addition of clamp A, the uncomplemented DNA (band ii) does not form any G4 structure, as evidenced by no apparent DMS protection pattern. Complemented DNA can either be in linear form or with clamp binding the flanking regions of alternate (e.g. not G41–4) G4 structures. As seen with band iii, a vague pattern of partial protection in multiple continuous guanine runs is evident, but a distinct G4 pattern is not noted. The suprashifted band iv has evident guanine protection in runs 1–4, confirming that not only is this highest band forming a G4 and dsDNA, but that it is particularly the target isoform: G41–4.
Clamp-mediated inhibition of promoter activity in vitro
Clamp a facilitates transcriptional downregulation
With the demonstration that clamps are capable of mediating G4 formation, and that they are highly selective for their target structure, we examined if any of the clamps were able to shift the DNA equilibrium in vitro and alter promoter activity. To do so, the MYC-containing Del4 plasmid (52) or the promoterless EV (pGL4.17) was transfected into HEK-293 cells alone or with one equivalence of each clamp for 24 or 48 h (Figure 6A). None of the clamps demonstrated any effect on promoter activity 24 h post-transfection. At 48 h post-transfection, only clamp A was able to significantly decrease promoter activity from the Del4 plasmid. Clamp C had no effect at any time point, and interestingly, clamp B facilitated a significant upregulation of MYC promoter activity 48 h post-transfection. A dose-response of clamp A at 48 h post transfection, a dose-dependent decrease in luciferase was noted with clamp A, which significantly decreased promoter activity at 0.5−5 equivalents (Figure 6B). Together, these works further support that G41–4, first noted in supercoiled DNA (51) and specifically recognized by clamp A, is the predominant physiological structure and that it is a transcriptional silencer of MYC.
Figure 6.
Clamp A downregulates MYC promoter activity. (A) Equimolar clamps A–C were co-transfected into HEK-293 cells along with the MYC-promoter driven firefly luciferase plasmid, Del4, and the control renilla plasmid, pRL. Cells were incubated for 24 or 48 h before lysing and measuring luciferase activity. MYC promoter activity was unaltered by any clamp after 24 h incubation, but was significantly decreased by clamp A 48 h post-transfection. Clamp B, at 48 h, significantly increased promoter activity; clamp C had no effect. (B) A dose-dependent effect of clamp A, transfected into HEK-293 cells as in (A), was examined 48 h post-transfection. Significantly decreased promoter activity was noted at doses as low as 0.5 eq, maintained through 5 equivalents. *p < 0.05 as compared to no clamp (0 eq) control, as determined by one-way ANOVA with post-hoc Tukey tests.
DISCUSSION
In the present work, we describe the development, optimization, and activity of a linked nucleic acid clamp capable of recognizing an individual G4 formed from the MYC NHE III1 promoter region. The clamps use an abasic polyethylene glycol phosphate linker to connect short nucleic acids complementing the 5′- and 3′-regions flanking the G4(s). The linker length was optimized to <18 Å, and the lead clamp (A) is able to specifically recognize its target isoform, G41–4. More than just recognizing the structure, clamp A demonstrated an ability to shift equilibrating DNA to enhance G4 formation, and that this affinity and specificity translates in vitro to an ability to decrease MYC promoter activity.
A number of equilibrating G4s have been described in the MYC promoter (55), most famously two unique structures with loops of 1, 2 and 1 nucleotide in the 5′-3′ direction, and a few studies highlighting a less frequent 1–6–1 loop isomer (G41–5) (54). From linear DNA, this 1:2:1 loop isomer structure forms from the second through the fifth run of continuous guanines as demonstrated by both DMS and NMR (G42–5) (52,53). Often it is this sequence that is used to screen for compounds or other nucleic acid-based therapeutics and to demonstrate their interaction. However, there is a notable study that demonstrates an alternate isoform occurring under supercoiled DNA conditions (51). This isoform is also a 1:2:1 loop isomer, but forms from the first through fourth runs of continuous guanines (G41–4). The effects of supercoiling are a major driving force in the transition from duplex to linear to G4 DNA, and thus its effects are quite physiologically relevant (28,56–58). The findings presented herein where clamp A, recognizing only this supercoiled structure, mediated a decrease in promoter activity supports G41–4 as the predominant, physiologically relevant structure in the MYC promoter. Supercoiling does not always shift the G4 structure from that found in linear DNA; the structure described for the VEGF promoter remains consistent (59–61); highlighting the critical need to examine each promoter individually and under conditions as physiologically relevant as possible.
In the same MYC supercoiling study, the i-motif (iM) formed on the C-rich strand was also described; it is noteworthy that this structure was also varied from the one identified to form from linear DNA. In addition, the G4 and iM structures formed under supercoiled conditions from the MYC promoter were offset from one another in such a manner that leaves a long string (14 nucleotides) of uncomplemented DNA on the 5′-side of the G4 (51). Computer modelling suggests that at least five nucleotides are needed to buffer the transition from higher order DNA structures to dsDNA (data not shown). The structures’ offset and the apparent required buffer region served as the basis for the length of the linked complementary bases in the clamps designed, and allows for a total length of complementary nucleotides of >20. Such a large number of complementing bases allows for great selectivity in binding across the genome and the linking of the regions fosters the specificity for structural recognition noted in the current study.
Clamp A's ability to modulate MYC promoter activity suggests it might have a therapeutic application. Most efforts to modulate G4 stability and MYC transcription are focused on the identification and generation of novel small molecules (41). With few exceptions (42,62–63), these molecules are often promiscuous or mediate their anti-cancer activity in a non-G4-mediated manner despite early evidence of G4 stabilization (64). This latter case may be, in part, due to the frequent use of G42–5 in screening efforts, versus G41–4. While these structures are similar, their loop constituent variations (T:GA:T versus A:TG:A, respectively) may prove critical to cellular activity. The first compound described to clearly mediate anti-cancer, and particularly anti-MYC, activity through the promoter G4 was NSC 338258, which was identified by screening with the entire NHE III1 region (42). Future efforts to develop small molecules against the MYC promoter G4 would likely be best served by either screening against the entire guanine-rich region, or with a G41–4 structure.
G4-targeted compounds often suffer from a generally low selectivity amongst similar structures (41), with a few exceptions (42,62–63). Non-small molecule based approaches to modulate MYC G4 formation have also been followed with greater success in target selectivity. In particular, the MYC NHE III1 region, in G4 formation, has been directly applied to leukemia cells. This structure was noted to have anti-leukemic activity related to modulation of telomere length and integrity and other non-telomeric related events (43,44). More specific to the MYC promoter and MYC G4's, peptide nucleic acid (PNA) probes have been designed. These PNA's can complement either the G- or the C-rich DNA strands (45,47,65) and have potential to inhibit dsDNA formation in the NHE III1 region and thus decrease promoter activity, although that is yet to be examined. The current work sought to build on the idea of the PNAs complementing the NHE III1, while maintaining G4 formation. While the complementing of DNA is fairly strong, it is a reversible process and thus will not damage DNA in a permanent manner as with PNA/DNA hydrids that mediate scission of the genome in conditions that are not present in nuclei; their physiological potential has not been explored (48). The clamps described are able to be specific for a particular promoter, in this case MYC, to facilitate non-canonical DNA formation, and to modulate transcription in a reversible manner.
Some recent work with strand invasion of the bcl-2 promoter further highlights the potential therapeutic potential of the clamps described in the present manuscript. This promoter contains four unique G4-forming regions with varying functions (66–69). One research group has taken the approach of PNA complementation the C-rich region of the most distal of the four G4-forming regions, enabling G4-formation and stabilization; invasion of the PNA is dependent on the ability to form the non-canonical structure, and not just sequence specific (68,70). A non-G4-related invading complementary, unmodified, oligonucleotide, PNT2258, has been developed to bind to a more-distal region of the bcl-2 promoter. This oligonucleotide decreases bcl-2 promoter activity, facilitates apoptosis, and is currently in phase II clinical trials as a liposomal package for diffuse large B-cell lymphoma (71,72). Albeit related to a different promoter and a non-G4-mechanism, this first-in-class DNA interference highlights the potential of targeting DNA and achieving clinical gain. The linked nucleic acid clamps described herein are a novel way to specifically target G4-controlled transcription. Future efforts with clamp A will be focused on optimizing its delivery to nuclei of cancer cells with nano-based polymers.
Acknowledgments
The authors wish to thank Vijay Gokhale for his expertise with computer modelling of higher order DNA structures.
Footnotes
Present address: Tracy A. Brooks, BioMolecular Sciences, University of Mississippi, University, MS 38677, USA.
FUNDING
National Institutes of Health [1P50CA13080501A1 subaward to T.A.B.]; University of Mississippi [to T.A.B.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dang C.V. Enigmatic MYC conducts an Unfolding Systems Biology Symphoma. Genes Cancer. 2010;1:526–531. doi: 10.1177/1947601910378742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudino T.A., McKay C., Pendeville-Samain H., Nilsson J.A., Maclean K.H., White E.L., Davis A.C., Ihle J.N., Cleveland J.L. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauters C., de Groote P., Adamantidis M., Delcayre C., Hamon M., Lablanche J.M., Bertrand M.E., Dupuis B., Swynghedauw B. Proto-oncogene expression in rabbit aorta after wall injury. First marker of the cellular process leading to restenosis after angioplasty. Eur. Heart J. 1992;13:556–559. doi: 10.1093/oxfordjournals.eurheartj.a060213. [DOI] [PubMed] [Google Scholar]
- 4.Biro S., Fu Y.M., Yu Z.X., Epstein S.E. Inhibitory effects of antisense oligodeoxynucleotides targeting c-myc mRNA on smooth muscle cell proliferation and migration. Proc. Natl. Acad. Sci. U.S.A. 1993;90:654–658. doi: 10.1073/pnas.90.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mai S., Fluri M., Siwarski D., Huppi K. Genomic instability in MycER-activated Rat1A-MycER cells. Chromosome Res. 1996;4:365–371. doi: 10.1007/BF02257272. [DOI] [PubMed] [Google Scholar]
- 6.Taylor C., Jalava A., Mai S. c-Myc dependent initiation of genomic instability during neoplastic transformation. Curr. Top. Microbiol. Immunol. 1997;224:201–207. doi: 10.1007/978-3-642-60801-8_20. [DOI] [PubMed] [Google Scholar]
- 7.Taylor C., Mai S. c-Myc-associated genomic instability of the dihydrofolate reductase locus in vivo. Cancer Detect. Prev. 1998;22:350–356. doi: 10.1046/j.1525-1500.1998.cdoa36.x. [DOI] [PubMed] [Google Scholar]
- 8.Yin X.Y., Grove L., Datta N.S., Long M.W., Prochownik E.V. C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene. 1999;18:1177–1184. doi: 10.1038/sj.onc.1202410. [DOI] [PubMed] [Google Scholar]
- 9.Crouch D.H., Fincham V.J., Frame M.C. Targeted proteolysis of the focal adhesion kinase pp125 FAK during c-MYC-induced apoptosis is suppressed by integrin signalling. Oncogene. 1996;12:2689–2696. [PubMed] [Google Scholar]
- 10.Gavioli R., Frisan T., Vertuani S., Bornkamm G.W., Masucci M.G. c-myc overexpression activates alternative pathways for intracellular proteolysis in lymphoma cells. Nat. Cell Biol. 2001;3:283–288. doi: 10.1038/35060076. [DOI] [PubMed] [Google Scholar]
- 11.Martin S.J., O'Brien G.A., Nishioka W.K., McGahon A.J., Mahboubi A., Saido T.C., Green D.R. Proteolysis of fodrin (non-erythroid spectrin) during apoptosis. J. Biol. Chem. 1995;270:6425–6428. doi: 10.1074/jbc.270.12.6425. [DOI] [PubMed] [Google Scholar]
- 12.O'Hagan R.C., Ohh M., David G., de Alboran I.M., Alt F.W., Kaelin W.G., DePinho R.A. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter F.E., Zucca E. Altered gene expression and oncogenesis of B-cell neoplasia. Ann. Oncol. 1991;2:335–342. doi: 10.1093/oxfordjournals.annonc.a057950. [DOI] [PubMed] [Google Scholar]
- 14.Kato G.J., Dang C.V. Function of the c-Myc oncoprotein. FASEB J. 1992;6:3065–3072. doi: 10.1096/fasebj.6.12.1521738. [DOI] [PubMed] [Google Scholar]
- 15.Lutz W., Leon J., Eilers M. Contributions of Myc to tumorigenesis. Biochim. Biophys. Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 16.Nesbit C.E., Tersak J.M., Prochownik E.V. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 17.Li B., Simon M.C. Molecular Pathways: Targeting MYC-induced metabolic reprogramming and oncogenic stress in cancer. Clin. Cancer Res. 2013;19:5835–5841. doi: 10.1158/1078-0432.CCR-12-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandopadhayay P., Bergthold G., Nguyen B., Schubert S., Gholamin S., Tang Y., Bolin S., Schumacher S.E., Zeid R., Masoud S., et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung M., Gelato K.A., Fernandez-Montalvan A., Siegel S., Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics. 2015;7:487–501. doi: 10.2217/epi.14.91. [DOI] [PubMed] [Google Scholar]
- 21.Sewastianik T., Prochorec-Sobieszek M., Chapuy B., Juszczynski P. MYC deregulation in lymphoid tumors: molecular mechanisms, clinical consequences and therapeutic implications. Biochim. Biophys. Acta. 2014;1846:457–467. doi: 10.1016/j.bbcan.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Shachaf C.M., Gentles A.J., Elchuri S., Sahoo D., Soen Y., Sharpe O., Perez O.D., Chang M., Mitchel D., Robinson W.H., et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res. 2008;68:5132–5142. doi: 10.1158/0008-5472.CAN-07-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shachaf C.M., Kopelman A.M., Arvanitis C., Karlsson A., Beer S., Mandl S., Bachmann M.H., Borowsky A.D., Ruebner B., Cardiff R.D., et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 24.Soucek L., Whitfield J., Martins C.P., Finch A.J., Murphy D.J., Sodir N.M., Karnezis A.N., Swigart L.B., Nasi S., Evan G.I. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamin L.R., Chung H.J., Sanford S., Kouzine F., Liu J., Levens D. Hierarchical mechanisms build the DNA-binding specificity of FUSE binding protein. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18296–18301. doi: 10.1073/pnas.0803279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levens D. How the c-myc promoter works and why it sometimes does not. J. Natl. Cancer Inst. Monogr. 2008:41–43. doi: 10.1093/jncimonographs/lgn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushita K., Tamura M., Tanaka N., Tomonaga T., Matsubara H., Shimada H., Levens D., He L., Liu J., Yoshida M., et al. Interactions between SAP155 and FUSE-binding protein-interacting repressor bridges c-Myc and P27Kip1 expression. Mol. Cancer Res. 2013;11:689–698. doi: 10.1158/1541-7786.MCR-12-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks T.A., Hurley L.H. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 29.Brooks T.A., Hurley L.H. Targeting MYC expression through G-quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev.: Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjostrom J., Blomqvist C., von Boguslawski K., Bengtsson N.O., Mjaaland I., Malmstrom P., Ostenstadt B., Wist E., Valvere V., Takayama S., et al. The predictive value of bcl-2, bax, bcl-xL, bag-1, fas, and fasL for chemotherapy response in advanced breast cancer. Clin. Cancer Res. 2002;8:811–816. [PubMed] [Google Scholar]
- 32.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLeod M.C. Identification of a DNA structural motif that includes the binding sites for Sp1, p53 and GA-binding protein. Nucleic Acids Res. 1993;21:1439–1447. doi: 10.1093/nar/21.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam E.Y., Beraldi D., Tannahill D., Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller S., Kumari S., Rodriguez R., Balasubramanian S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010;2:1095–1098. doi: 10.1038/nchem.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decorsière A., Cayrel A., Vagner S., Millevoi S. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari S., Bugaut A., Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcel V., Tran P.L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M.P., Hall J., Mergny J.L., Hainaut P., Van Dyck E. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 41.Morgan R.K.B.T.A. In: Small-molecule Transcription Factor Inhibitors in Oncology. Rahman MT, Thurston D, editors. RSC Drug Discovery; 2016. [Google Scholar]
- 42.Brown R.V., Danford F.L., Gokhale V., Hurley L.H., Brooks T.A. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J. Biol. Chem. 2011;286:41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Islam M.A., Thomas S.D., Murty V.V., Sedoris K.J., Miller D.M. c-Myc quadruplex-forming sequence Pu-27 induces extensive damage in both telomeric and nontelomeric regions of DNA. J. Biol. Chem. 2014;289:8521–8531. doi: 10.1074/jbc.M113.505073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sedoris K.C., Thomas S.D., Clarkson C.R., Muench D., Islam A., Singh R., Miller D.M. Genomic c-Myc quadruplex DNA selectively kills leukemia. Mol. Cancer Ther. 2012;11:66–76. doi: 10.1158/1535-7163.MCT-11-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A., Lee L.L., Roy S., Tanious F.A., Wilson W.D., Ly D.H., Armitage B.A. Strand invasion of DNA quadruplexes by PNA: comparison of homologous and complementary hybridization. Chembiochem. 2013;14:1476–1484. doi: 10.1002/cbic.201300263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar N., Patowary A., Sivasubbu S., Petersen M., Maiti S. Silencing c-MYC expression by targeting quadruplex in P1 promoter using locked nucleic acid trap. Biochemistry. 2008;47:13179–13188. doi: 10.1021/bi801064j. [DOI] [PubMed] [Google Scholar]
- 47.Roy S., Tanious F.A., Wilson W.D., Ly D.H., Armitage B.A. High-affinity homologous peptide nucleic acid probes for targeting a quadruplex-forming sequence from a MYC promoter element. Biochemistry. 2007;46:10433–10443. doi: 10.1021/bi700854r. [DOI] [PubMed] [Google Scholar]
- 48.Ishizuka T., Yang J., Komiyama M., Xu Y. G-rich sequence-specific recognition and scission of human genome by PNA/DNA hybrid G-quadruplex formation. Angew. Chem. 2012;51:7198–7202. doi: 10.1002/anie.201201176. [DOI] [PubMed] [Google Scholar]
- 49.Lavrado J., Brito H., Borralho P.M., Ohnmacht S.A., Kim N.S., Leitao C., Pisco S., Gunaratnam M., Rodrigues C.M., Moreira R., et al. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci. Rep. 2015;5:9696. doi: 10.1038/srep09696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 51.Sun D., Hurley L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 54.Dettler J.M., Buscaglia R., Le V.H., Lewis E.A. DSC deconvolution of the structural complexity of c-MYC P1 promoter G-quadruplexes. Biophys. J. 2011;100:1517–1525. doi: 10.1016/j.bpj.2011.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le H.T., Miller M.C., Buscaglia R., Dean W.L., Holt P.A., Chaires J.B., Trent J.O. Not all G-quadruplexes are created equally: an investigation of the structural polymorphism of the c-Myc G-quadruplex-forming sequence and its interaction with the porphyrin TMPyP4. Org. Biomol. Chem. 2012;10:9393–9404. doi: 10.1039/c2ob26504d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 57.Travers A., Muskhelishvili G. DNA supercoiling—a global transcriptional regulator for enterobacterial growth. Nat. Rev.: Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 58.Wang Z., Droge P. Differential control of transcription-induced and overall DNA supercoiling by eukaryotic topoisomerases in vitro. EMBO J. 1996;15:581–589. [PMC free article] [PubMed] [Google Scholar]
- 59.Sun D., Guo K., Rusche J.J., Hurley L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun D., Guo K., Shin Y.J. Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Res. 2011;39:1256–1265. doi: 10.1093/nar/gkq926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun D., Liu W.J., Guo K., Rusche J.J., Ebbinghaus S., Gokhale V., Hurley L.H. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Felsenstein K.M., Saunders L.B., Simmons J.K., Leon E., Calabrese D.R., Zhang S., Michalowski A., Gareiss P., Mock B.A., Schneekloth J.S., Jr Small molecule microarrays enable the identification of a selective, quadruplex-binding inhibitor of MYC Expression. ACS Chem. Biol. 2016;11:139–148. doi: 10.1021/acschembio.5b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaiser C.E., Gokhale V., Yang D., Hurley L.H. Gaining insights into the small molecule targeting of the G-quadruplex in the c-MYC promoter using NMR and an allele-specific transcriptional assay. Top. Curr. Chem. 2013;330:1–21. doi: 10.1007/128_2012_333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boddupally P.V., Hahn S., Beman C., De B., Brooks T.A., Gokhale V., Hurley L.H. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J. Med. Chem. 2012;55:6076–6086. doi: 10.1021/jm300282c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohammed H.S., Delos Santos J.O., Armitage B.A. Noncovalent binding and fluorogenic response of cyanine dyes to DNA homoquadruplex and PNA-DNA heteroquadruplex structures. Artif. DNA PNA XNA. 2011;2:43–49. doi: 10.4161/adna.2.2.16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dexheimer T.S., Sun D., Hurley L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onel B., Carver M., Wu G., Timonina D., Kalarn S., Larriva M., Yang D. A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc. 2016;138:2563–2570. doi: 10.1021/jacs.5b08596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Onyshchenko M.I., Gaynutdinov T.I., Englund E.A., Appella D.H., Neumann R.D., Panyutin I.G. Stabilization of G-quadruplex in the BCL2 promoter region in double-stranded DNA by invading short PNAs. Nucleic Acids Res. 2009;37:7570–7580. doi: 10.1093/nar/gkp840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun H., Xiang J., Shi Y., Yang Q., Guan A., Li Q., Yu L., Shang Q., Zhang H., Tang Y., et al. A newly identified G-quadruplex as a potential target regulating Bcl-2 expression. Biochim. Biophys. Acta. 2014;1840:3052–3057. doi: 10.1016/j.bbagen.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 70.Onyshchenko M.I., Gaynutdinov T.I., Englund E.A., Appella D.H., Neumann R.D., Panyutin I.G. Quadruplex formation is necessary for stable PNA invasion into duplex DNA of BCL2 promoter region. Nucleic Acids Res. 2011;39:7114–7123. doi: 10.1093/nar/gkr259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shukkur Ebrahim A., Kandouz M., Liddane A., Sabbagh H., Hou Y., Li C., Al-Katib A. PNT2258, a novel deoxyribonucleic acid inhibitor, induces cell cycle arrest and apoptosis via a distinct mechanism of action: a new class of drug for non-Hodgkin's lymphoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9872. doi:10.18632/oncotarget.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tolcher A.W., Rodrigueza W.V., Rasco D.W., Patnaik A., Papadopoulos K.P., Amaya A., Moore T.D., Gaylor S.K., Bisgaier C.L., Sooch M.P., et al. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014;73:363–371. doi: 10.1007/s00280-013-2361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]