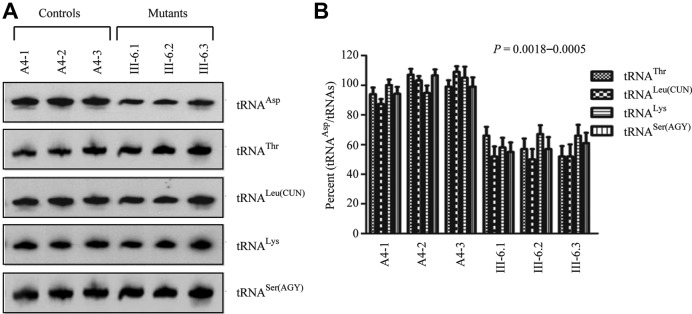
Figure 2.
Northern blot analysis of mitochondrial tRNA. (A) Equal amounts of total mitochondrial RNA from various cell lines were electrophoresed through a denaturing polyacrylamide gel, electroblotted and hybridized with DIG-labeled oligonucleotide probes specific for the mt–tRNAAsp, mt–tRNAThr, mt–tRNALeu(CUN), mt–tRNALys and mt–tRNASer(AGY), respectively. (B) Quantification of tRNA levels. Average relative mt–tRNAAsp content per cell, was normalized to the average content per cell of mt–tRNAThr, mt–tRNALeu(CUN), mt–tRNALys and mt–tRNASer(AGY) in three cybrid cell lines derived from one affected subject (III-6) carrying the m.7551A > G mutation and three cybrid cell lines derived from one Chinese control subject (A4). The values for the latter are expressed as percentages of the average values for the control cell lines. The calculations were based on three independent determinations of each tRNA content in each cell line and three determinations of the content of reference tRNA marker in each cell line. The error bars indicate two standard errors of the means. P indicates the significance, according to the t-test, of the differences between mutant and control cell lines.