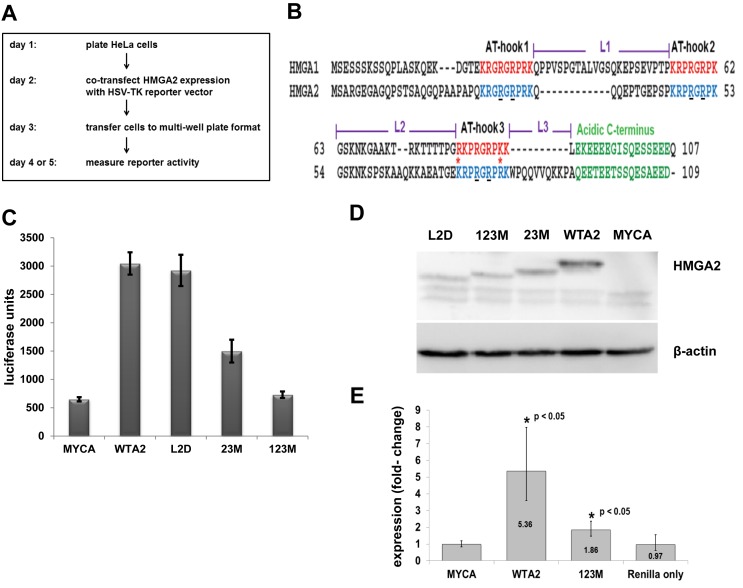
Figure 1.
Transcriptional activation of HSV-TK promoter depends on functional AT-hooks. (A) Diagram showing the workflow of the reporter system. (B) Sequence alignment of human HMGA1 and HMGA2 proteins, with AT-hooks, linker (L) and C-terminal domains highlighted. Asterisks indicate sequence differences in AT-hook 3 between HMGA1 and HMGA2 (adapted from (9)). The underlined R residues in HMGA2 AT-hooks demarcate glycine substitutions in mutated HMGA2, as described in the text. (C) Representative luciferase reporter assays of co-transfections comparing mock (MYCA), wild-type HMGA2 (WTA2), two AT-hook mutants (23M, 123M) and linker 2 deletion mutant (L2D). We show mean values plus standard deviations of readings from eight replicates per co-transfected plasmids. (D) Western blot analysis of expression of HMGA2 proteins using anti-FLAG antibody and β-actin as loading control. Note that the HMGA2 mutants exhibit a different electrophoretic mobility from the wild-type protein. (E) qRT-PCR analysis of Renilla mRNA after co-transfection with wild type or mutant HMGA2-expression constructs. We used the ΔΔCT method to determine the expression changes from four independent sets of co-transfections, each comparing mock, wild-type HMGA2, mutant HMGA2 and solely transfected Renilla expression vector. Data were normalized to GAPDH mRNA levels. Error bars, mean ± standard deviation. Statistical significance compared to mock transfections as indicated, determined using one-way ANOVA analysis. Nearly identical results were obtained when data were analyzed with Pfaffl's method (not shown).