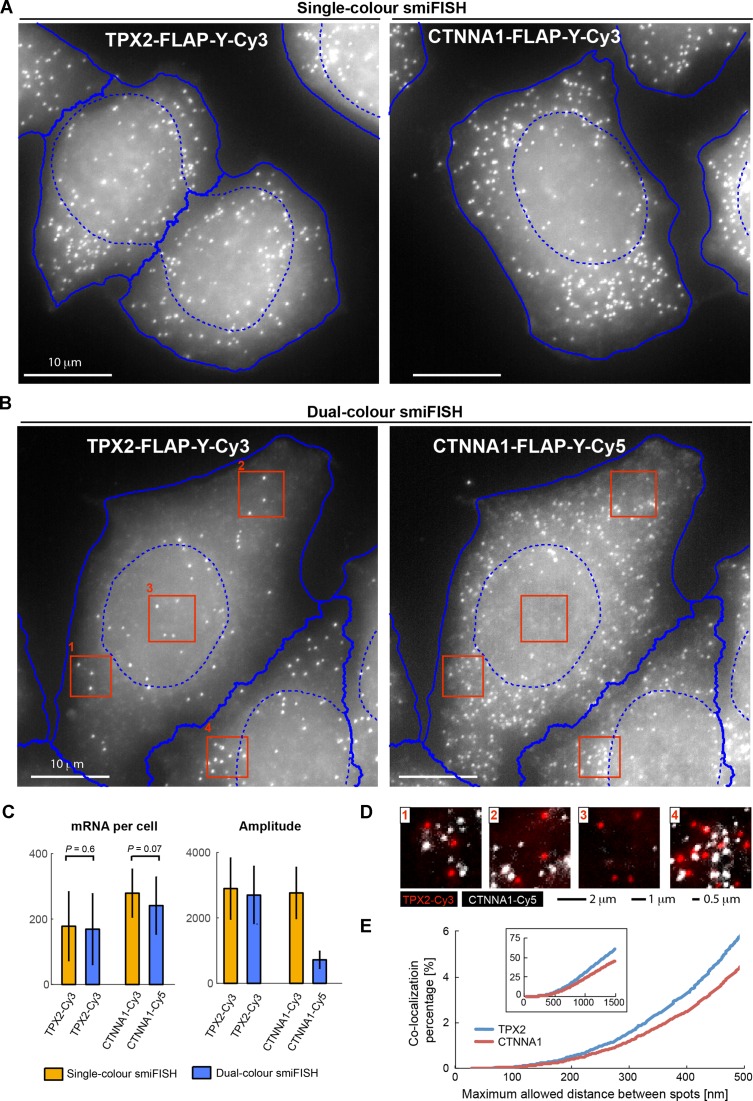
Figure 3.
Detection of CTNNA1 and TPX2 mRNA by two-colour smiFISH in HeLa cells. Single-colour smiFISH performed with Cy3 against either gene. Dual-colour smiFISH performed with Cy3 against TPX2, and Cy5 against CTNNA1. Both probe-sets contain FLAP-Y, and were prehybridized separately with the secondary probes before the actual experiment. (A and B) Maximum intensity projections of representative smiFISH images for (A) single-colour and (B) dual-colour experiments. For automated segmentation, nuclei were marked with DAPI, and cells with HCS CellMaskTM Green (Molecular Probes). Segmentation was performed on focus-projected images (see last section of results) with CellCognition (10): nuclei shown with dashed blue lines, cells with solid blue lines. (C) Left: estimated mRNA number per cell. Reported P-value from Wilcoxon rank sum test for equal medians (Matlab function ranksum). Right: amplitude of 3D Gaussian fit to each detected mRNA molecule. TPX2 mRNA detection was similar in single-colour and dual-colour experiments with respect to estimated mRNA counts and mRNA spot intensity. Labelling of CTNNA1 mRNA with Cy5 leads to substantially dimmer spots, which makes mRNA detection more challenging and leads to a lower P-value for the comparison of mRNA counts. Number of cells per condition: NCTTNA1 = 143, NTPX2 = 115; NCTTNA1,TPX2 = 150. (D) Dual-colour smiFISH (TPX2 in red, and CTNNA1 in white). Images are zoom-ins indicated by red rectangles in (B). For better visualization, images were background corrected with the Subtract Background function in Fiji (Rolling ball radius of 50 pixels). (E) Co-localization analysis for dual-colour smiFISH experiment in (C). mRNAs were detected in 3D images with FISH-quant (5) and co-localization determined with the Matlab function munkres using the Hungarian Algorithm for linear assignment problems, which is available on Matlab File Exchange. Plots show the co-localization percentage for TPX2 (blue, 27 178 mRNAs) and CTNNA1 (red, 35 623 mRNAs) as a function of the maximum allowed distance between spots to be still considered to be co-localized. For distances smaller than 500 nm, co-localization is smaller than 5%. Larger allowed distances lead to a significant increase of co-localization percentage, since neighbouring spots are erroneously considered to co-localize (especially in denser area such as zoom-in 4).