Figure 1.
Schematic illustrations of the non-cooperative and cooperative binding of gp32 monomers to: (A) the 3′-tail-pdT7-Cy3/Cy5 p/t DNA construct; (B) the 3′-tail-pdT15-Cy3/Cy5 p/t DNA construct; and (C) the 3′-tail-pdT22-Cy3/Cy5 p/t DNA construct. The single-stranded regions of these DNA constructs can bind (A) 0 or 1, (B) 0, 1 or 2 or (C) 0, 1, 2 or 3 gp32 proteins, respectively. (D) Bulk fluorescence measurements of the 3′-tail-pdT15-Cy3/Cy5 construct at 100 nM oligomer concentration exhibited changes in the FRET efficiency upon titration with gp32. Red, blue and green curves are partially overlapping. The inset shows the values of 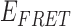 calculated from the peak Cy3/Cy5 fluorescence intensities. (E) The results of bulk fluorescence gp32 titration measurements (at 100 mM NaCl, 6 mM MgCl2 and 10 mM Tris (pH 8.0) concentrations) for initial 100 nM concentrations of six of the p/t DNA constructs used in our studies (for both 5′ and 3′ oliogo-dT tails of length N = 7, 15 and 22).
calculated from the peak Cy3/Cy5 fluorescence intensities. (E) The results of bulk fluorescence gp32 titration measurements (at 100 mM NaCl, 6 mM MgCl2 and 10 mM Tris (pH 8.0) concentrations) for initial 100 nM concentrations of six of the p/t DNA constructs used in our studies (for both 5′ and 3′ oliogo-dT tails of length N = 7, 15 and 22).