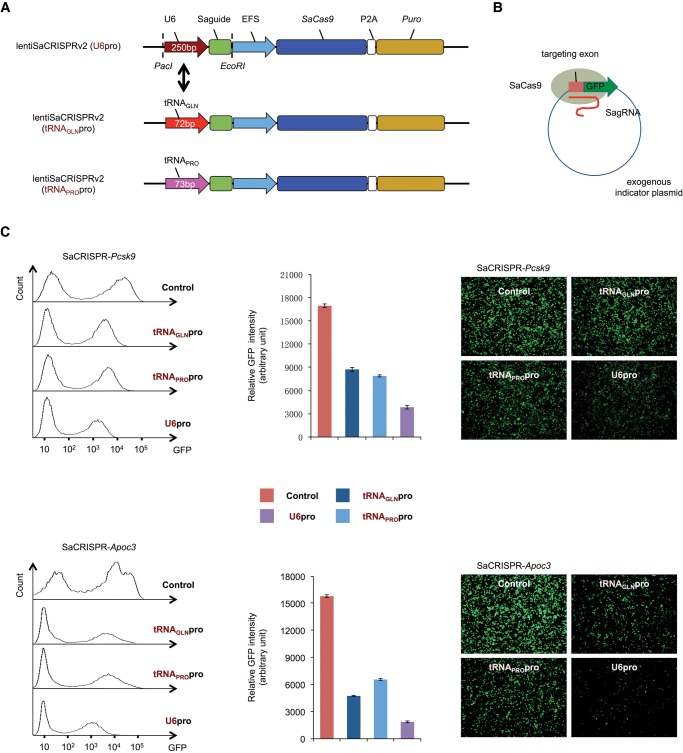
FIGURE 1.
Study of editing efficiency of CRISPR/Cas9 with tRNA and U6 promoter-driven sgRNAs by using exogenous indicator plasmids. (A) LentiSaCRISPR v2 was modified from lentiCRISPR v2 by switching the respected Cas9 and sgRNA cassettes. tRNAGLN(GGTTCCATGGTGTAATGGTTAGCACTCTGGACTCTGAATCCAGCGATCCGAGTTCAAATCTCGGTGGAACCT) and tRNAPRO(TGGCTCGTTGGTCTAGGGGTATGATTCTCGCTTCGGGTGCGAGAGGTCCCGGGTTCAAATCCCGGACGAGCCC) promoter-driven sgRNA cassettes were synthesized and inserted between PacI and EcoRI cutting sites. (B) A schematic diagram of an editing efficiency study by using exogenous indicator plasmids. (C) gRNAs targeting endogenous mouse Pcsk9 and Apoc3 genes were cloned into a lentiSaCRISPR v2 plasmid. An extra G was added for Pcsk9 and Apoc3 targeting when the U6 promoter was used. Indicator plasmids were constructed by inserting sequences of targeted exons to the pEGFP-N1 plasmid between two restriction enzyme sites at the MCS region and 5′ to the GFP gene. Different combinations of lentiSaCRISPR and indicator plasmids were cotransfected to HEK 293T cells at a mass ratio of 3:1 as indicated, and cells were harvested for FACS analysis and GFP imaging after 72 h. Three independent experiments were carried out, and representative images from one experiment are shown. Average of GFP mean intensity in FACS analysis from all three experiments with SEM is indicated. The gRNA sequences are listed in Supplemental Table 1.