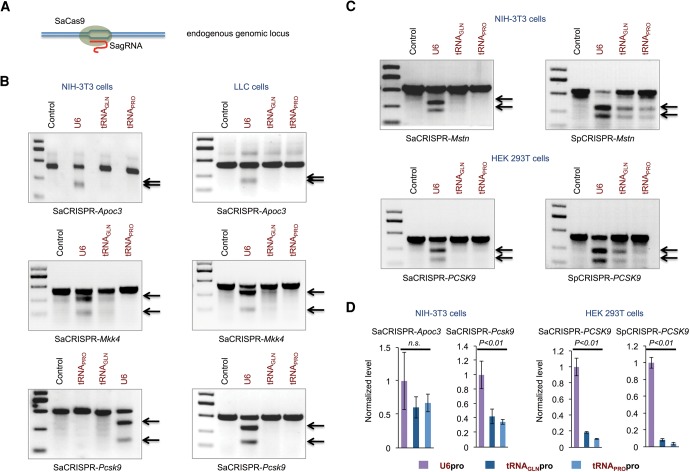
FIGURE 2.
Study of genome editing efficiency of CRISPR/Cas9 with tRNA and U6 promoter-driven sgRNAs by directly accessing mutagenesis rate at the endogenous genomic loci. (A) A schematic diagram of editing efficiency study by directly accessing the endogenous genomic loci. (B) Viral vectors targeting endogenous mouse Apoc3, Mkk4, and Pcsk9 genes were produced in HEK 293T cells with different lentiSaCRISPR constructs, and mouse NIH-3T3 cells and LLC cells were infected and selected with puromycin to enrich for cells transduced with viruses. Cells were then harvested for genomic DNA extraction and Surveyor assays for editing efficiency studies. (C) Viral vectors targeting endogenous mouse Mstn gene and human PCSK9 gene were produced with both lentiSaCRISPR and lentiSpCRISPR constructs, and mouse NIH-3T3 cells (for Mstn targeting) and human HEK 293T cells (for PCSK9 targeting) were infected, selected with puromycin and harvested for Surveyor assays. The gRNA sequences as well as sequences of primers used in Surveyor assays in B and C are listed in Supplemental Table 1. Arrows show the cleavage products resulting from the Surveyor assays; the intensity of the cleavage product bands relative to the uncleaved product band corresponds to the mutagenesis rate. (D) Cells infected with viruses as indicated were collected and subjected to small-RNA extraction. Poly(A) tails were added to RNA before reverse transcription was performed. The expression level of sgRNAs was evaluated with quantitative PCR using SYBR green. 18S RNA level was used as an internal control. The sequences of primers for qRT-PCR were listed in Supplemental Table 1. All data are represented as means with SEM. One-way ANOVA was used for statistical analysis.