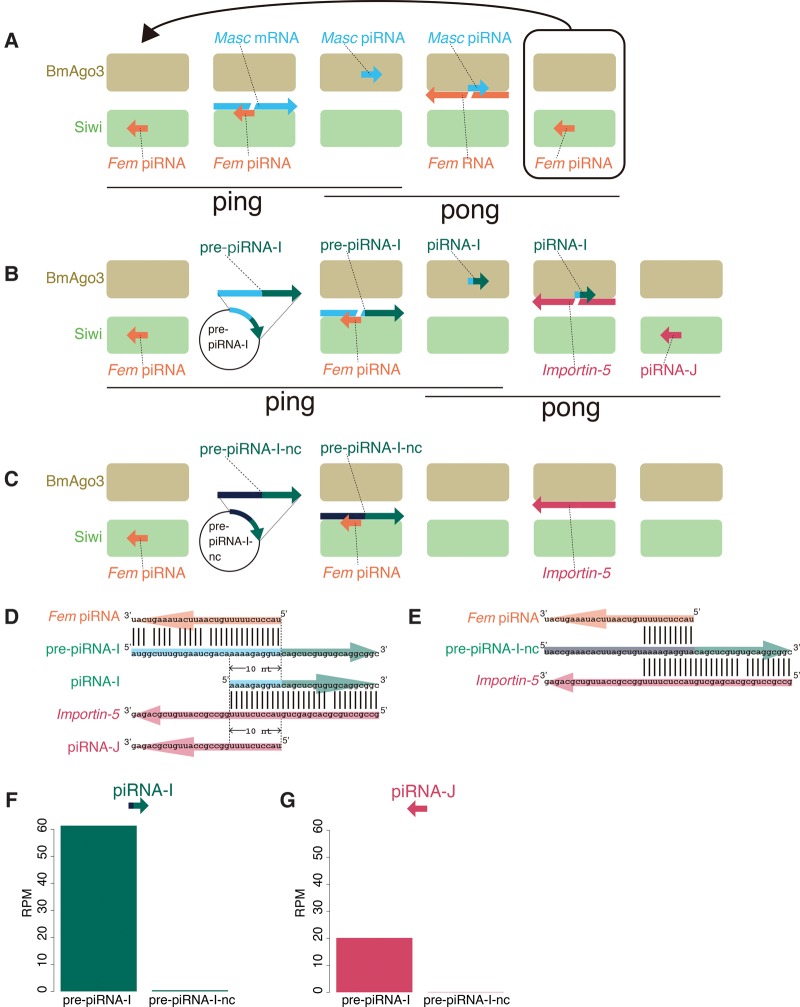
FIGURE 1.
Conceptual diagrams of the artificial piRNA production system using silkworm BmN-4 cells. (A) The silkworm sex determination pathway via a unique one-to-one ping-pong pair, Fem piRNA and Masc piRNA. (B,C) The artificial piRNA production system targeting the silkworm Importin-5 gene. The silkworm ovary-derived cell line BmN-4 was transfected with plasmids expressing pre-piRNA-I (B) or pre-piRNA-I-nc (C). pre-piRNA-I mRNA is cleaved by the Fem piRNA–Siwi complex, whereas pre-piRNA-I-nc mRNA is not cleaved, due to sequence incompatibility. The resulting 3′ pre-piRNA-I fragment is loaded into BmAgo3, producing the piRNA-I–BmAgo3 complex, which recognizes and cleaves silkworm Importin-5 mRNA. The cleaved 3′ fragment is then loaded into another PIWI protein, Siwi, thereby producing the piRNA-J–Siwi complex. (D,E) Sequence design for artificial piRNA precursors. The pre-piRNA-I is designed to be cleaved by the endogenous Fem piRNA–Siwi complex, whereas pre-piRNA-I–nc mRNA contains nontargeted nucleotide sequences. (F,G) Normalized piRNA-I (F) and piRNA-J (G) reads in total piRNA libraries from BmN-4 cells transfected with pre-piRNA-expressing plasmids. Mapping reads against 1811 transposons were used for normalization.