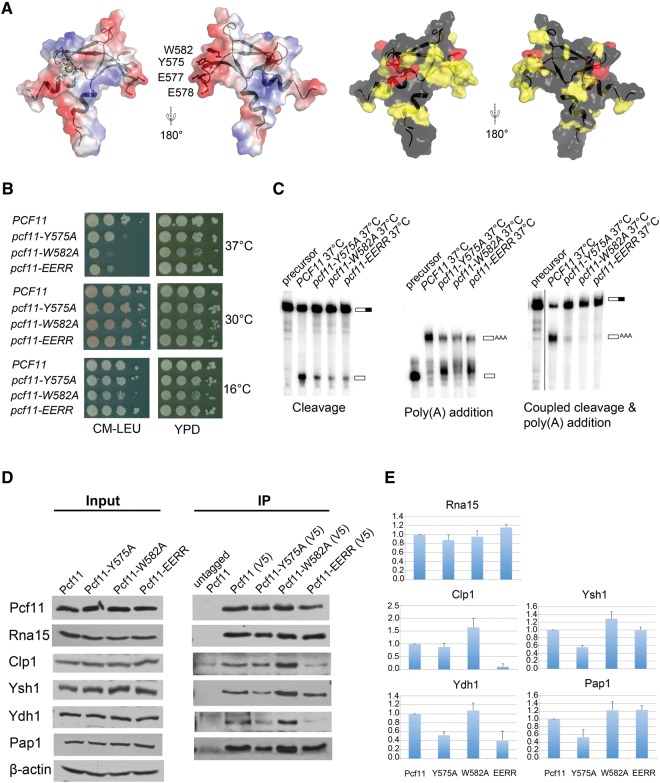
FIGURE 4.
Pcf11 is necessary for both cleavage and polyadenylation steps of mRNA 3′-end processing in yeast. (A) Highly conserved residues cluster in a negative-charged binding surface. The left panel corresponds to surface representations showing the electrostatic potential, with red indicating negative and blue indicating positive electrostatic potential, with the four mutated residues indicated. The right panel shows surface representations of conserved residues (completely conserved in red, less conserved in yellow). (B) Growth characteristics of pcf11-W582A, pcf11-Y575A, and pcf11-EERR mutant strains at 16°C, 30°C, and 37°C. (C) In vitro cleavage assay (left), in vitro polyadenylation assay (middle), and coupled cleavage and polyadenylation assay (right). The experiments were carried out at 37°C. The positions of precursor and product RNAs are indicated on the right side of each panel. (D) Coimmunoprecipitation with extracts prepared from cells expressing forms of Pcf11, Pcf11-Y575A, Pcf11-W582A, and Pcf11-EERR tagged with both V5 and Myc. Cells were grown at 30°C, then shifted to 37°C for 2 h. Coimmunoprecipitation was performed using anti-V5 antibody and the eluates were analyzed by Western blotting with antibodies against CF IA and CPF subunits. Pcf11 was detected with the Myc antibody. The control was done with extract from cells expressing only untagged Pcf11. The left panel shows the input used for each extract, with β-actin as the loading control. (E) Quantitation of co-IP analysis. The ratio of coimmunoprecipitated protein to that of Pcf11 was determined by scanning the Western blots using ImageJ for pulldowns from two different extract preparations. These ratios were then divided by the ratios observed for the wild-type protein and plotted to generate relative binding activities.