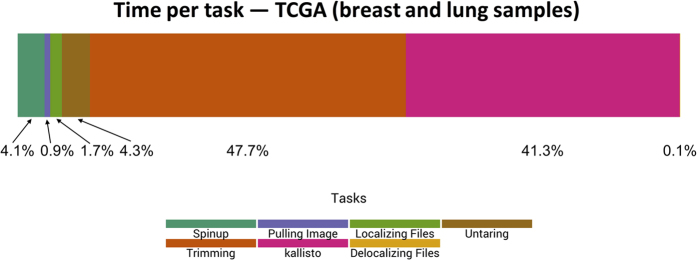
Figure 4. Relative time spent on computational tasks for TCGA breast and lung samples using a preemptible-node configuration.
We logged the durations of individual processing tasks for the TCGA breast and lung samples, averaged these values, and calculated the percentage of overall processing time for each task. The “spinup,” image pulling, and file localization steps enabled the virtual machines to begin executing. For sample preprocessing, the FASTQ files were unpacked, decompressed, and quality trimmed; together these steps took 52.0% of the processing time (on average). The kallisto alignment and quantification steps took 41.3% of the overall processing time.