Figure 2.
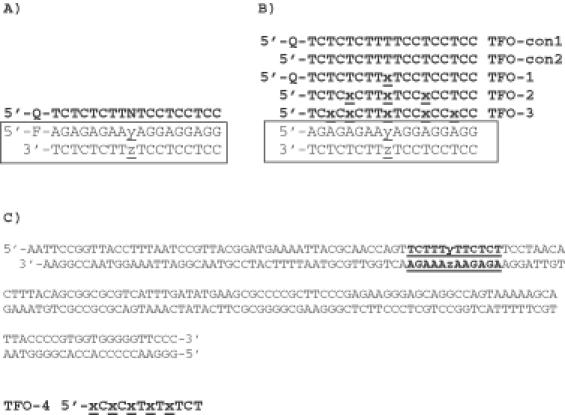
Sequence of oligonucleotides used in this work. In each case the third strand is shown in bold and the duplex target is boxed. (A) Oligonucleotides used for fluorescence melting experiments, Q, methyl red serinol; F, 6-amidohexylfluorescein; N, BAU T, A, G or C; y.z = each base pair in turn TA, AT, CG or GC. (B) Oligonucleotides used for UV melting studies x, BAU; y.z, each base pair in turn TA, AT, CG, or GC. TFO-1 (containing one BAU residue) was identical to one of the oligonucleotides used for the fluorescence melting studies. (C) Sequence of the footprinting substrates derived from tyrT(43–59); y.z = each base pair in turn in different fragments. The DNA was labelled at the 3′ end of the EcoRI site (lower strand). The triplex target site is underlined and in bold and was targeted with TFO-4 (x = BAU).
