Figure 4.
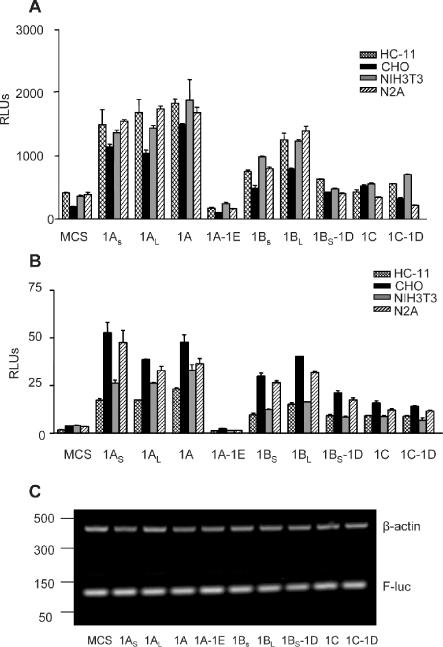
Varying translational efficiencies of the different cx43 5′-UTRs. (A) Relative luciferase (Firefly/Renilla) activities of various cell lines transiently transfected with 1.2 μg of pcDNA3.1 (Stratagene)-derived constructs containing each of the mouse cx43 5′-UTRs sequences upstream of the luciferase coding region. MCS correspond to the relative luciferase values obtained with the empty pcDNA3.1(+) vector. Cells were co-transfected with 15 ng of the pRL-TK Renilla expressing vector (Promega) to correct for variations in transfection efficiency. Values are expressed as the means ± SE of at least three replicate experiments. The cell lines included HC-11, CHO, NIH3T3 and N2A cells. (B) As in (A), but cells were co-transfected with 1 μg of in vitro transcribed, capped, chimeric cx43 5′-UTR/luciferase reporter mRNAs and 50 ng of Renilla-luciferase mRNA, as indicated. MCS corresponds to the relative luciferase values obtained from the in vitro transcribed luciferase mRNA from pSPluc+[A]60 vector. (C) Semiquatitative RT–PCR analysis of total RNA obtained from N2A cells transfected with in vitro transcribed, capped, either chimeric cx43 5′-UTR/luciferase reporter mRNAs or mRNA transcribed from the empty pSPluc+[A]60 vector (MCS), 6 h after transfection. The RT–PCR reactions contained one set of primers annealing to the Firefly-luciferase coding region (F-luc), and a second set of primers annealing to the β-actin gene (see Materials and Methods for primer sequences). The PCR-amplified products were separated in a 2% agarose gel and visualized by ethidium bromide staining. The positions of the molecular weight markers (PCR markers; Promega) are indicated on the left-hand side.
