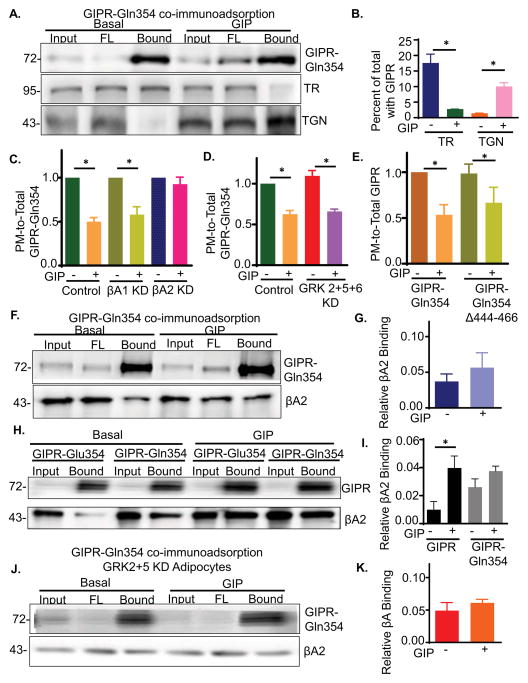
Fig. 6. Redistribution of the GIPR-Gln354 variant to the TGN is dependent on GIP and βA2 and independent of GRKs.
(A) HA-GIPR-Gln354-GFP Immunoadsorption from WT adipocytes and blotted for GIPR, TR and TGN46.
(B) Quantification of data from experiments like that shown in panel A. TR and TGN46 in elution were calculated as normalized in total (input) and GIPR in the elution. Data are average ± SD from 3 independent experiments.
(C) Effect βA1 or βA2 KD on PM-to-Total distribution of GIPR-Gln354 variant. Data are normalized to their own basal values. Data are averages ± SD of 3 independent experiments., P<0.05.
(D) Effect of GRK2+5+6 triple KD on PM-to-Total distribution of GIPR-Gln354 variant. Data are averages ± SD of 3 independent experiments., P<0.05.
(E) PM-to-Total distribution of GIPR-Gln354 variant and GIPR-354Gln-Δ444-466 deletion mutant. Data are averages ± SD of 3 independent experiments., P<0.05.
(F) Co-immunoprecipitation of βA2 with GIPR-Gln354.
(G) Quantification of data from experiments like that shown in panel F. Data are average ± SD from 3 independent experiments.
(H) Relative βA2 binding to GIPR and GIPR-354Gln. GIPR and GIPR-354Gln Co-IP were blotted for GIPR and βA2 on the same blot.
(I) Quantification of data from experiments like that shown in panel H. Data are average ± SD from 3 independent experiments.
(J) GIPR-354Gln-GFP:βA2 binding in GRK2+5 double KD cells.
(K) Quantification of data from experiments like that shown in panel H. Data are average ± SD from 3 independent experiments.