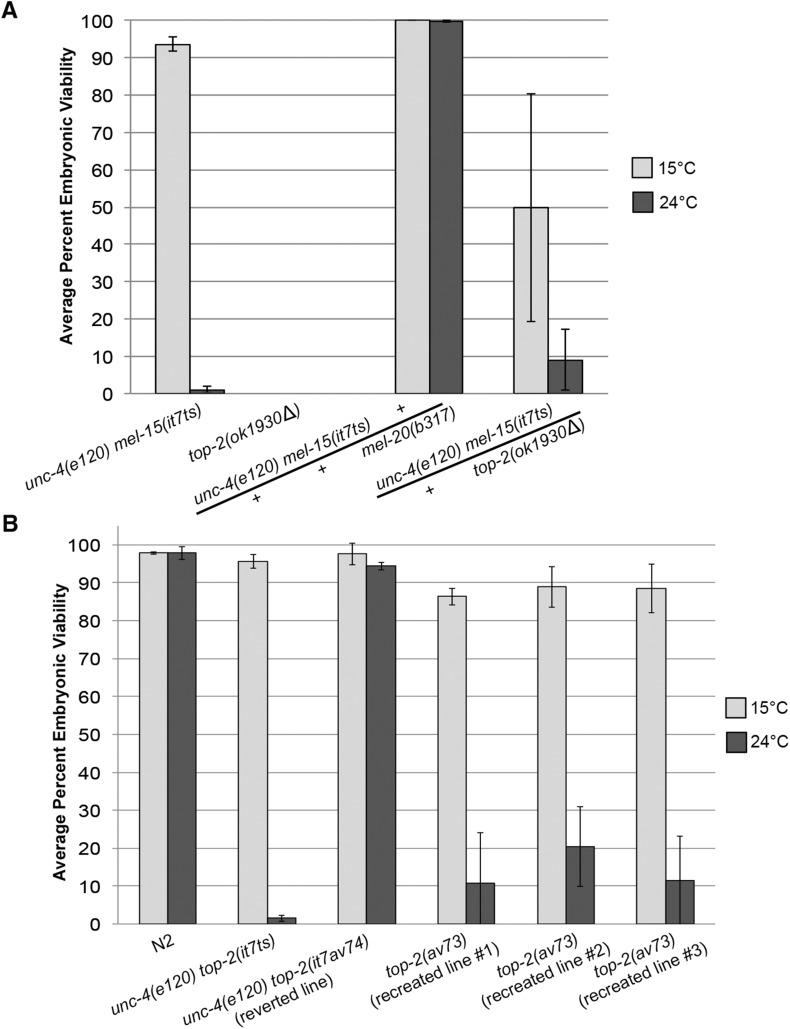
Figure 2.
mel-15(it7ts) paternal-effect embryonic lethality is caused by a point mutation in top-2. (A) Complementation tests were performed by crossing unc-4(e120) mel-15(it7ts) hermaphrodites with either top-2(ok1930Δ)/mIn1-gfp; him-8(e1489) or mel-20(b317)/mnC1-gfp; him-8(e1489) males. The average percent embryonic viability at 15 or 24° was calculated by scoring the progeny of F1 trans-heterozygous hermaphrodites. The number of hermaphrodites scored for each cross at 15° were: unc-4(e120) mel-15(it7ts)/mel-20(b317), n = 30; and unc-4(e120) mel-15(it7ts)/top-2(ok1930Δ), n = 42. The number of hermaphrodites scored for each cross at 24° were unc-4(e120) mel-15(it7ts)/mel-20(b317), n = 30; and unc-4(e120) mel-15(it7ts)/top-2(ok1930Δ), n = 40. The average percent embryonic viability for self-mating unc-4(e120) mel-15(it7ts) and top-2(ok1930Δ) hermaphrodites is also indicated (15°, n = 36 and 16, respectively; 24°, n = 21 and 20). Note: top-2(ok1930Δ) homozygotes are sterile Uncs and do not produce any progeny. Error bars indicate SD. (B) Average percent embryonic viability from N2 (wild type), unc-4(e120) top-2(it7ts), and the CRISPR/Cas9-mediated reverted line of unc-4(e120) top-2(it7ts) to wild type [unc-4(e120) top-2(av74)], and the three it7ts recreated lines in N2 [top-2(av73)] at 15 or 24°. The progeny of at least 28 hermaphrodites were scored for each strain at 15 and 24°. Error bars represent SD of the average percentage of three individual replicate experiments.