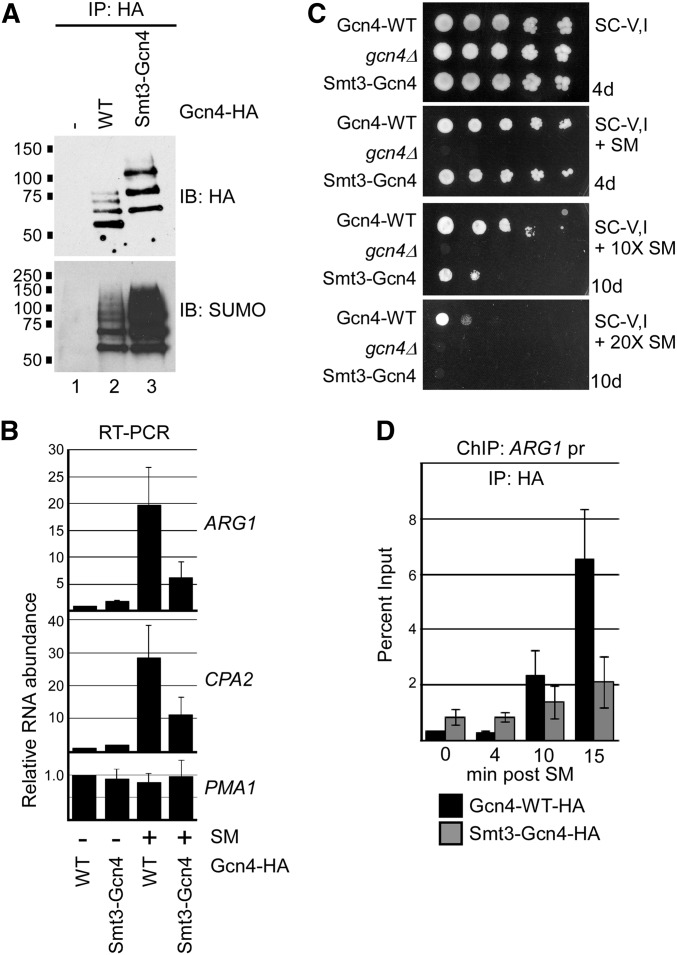
Figure 5.
Hyper-sumoylation of Gcn4 reduces its occupancy on target DNA. (A) HA and SUMO immunoblot analysis of HA IPs of Gcn4-WT, or of the fusion Smt3-Gcn4-WT, both of which contain the usual C-terminal 6× HA tag. A strain with no HA tag on Gcn4 (−) was analyzed in parallel as a control. Strains analyzed are ERYM615, ERYM613, and YAA030H. (B) qRT-PCR analysis of RNA isolated from Gcn4-WT or Smt3-Gcn4 strains mock-treated (−) or induced for Gcn4 expression with SM (+). Genes analyzed include Gcn4-targets ARG1 and CPA2, and the constitutively expressed PMA1 gene. The average of three independent experiments is shown, with SD shown as error bars. Strains analyzed are ERYM613 and YAA030H. (C) Yeast spot assay comparing growth of strains expressing Gcn4-WT, no Gcn4 (gcn4Δ), or the fusion Smt3-Gcn4 on minimal medium lacking Val and Ile, and supplemented with either no SM (top), 0.5 μg/ml SM, or 10× or 20× this concentration, as indicated. Plates were photographed after indicated number of days of growth. Strains analyzed are ERYM613, ERYM625, and YAA030H. (D) Comparison of Gcn4 and Smt3-Gcn4 occupancy on target DNA. HA ChIP analysis of the promoter-proximal region of the ARG1 gene was performed in the Gcn4-WT or Smt3-Gcn4 strains at indicated times post induction with SM. The average of three independent experiments is shown with SD shown as error bars. Strains analyzed are ERYM613 and YAA030H.