Abstract
DNA flap endonuclease 1 (FEN1) plays critical roles in maintaining genome stability and integrity by participating in both DNA replication and repair. Suppression of FEN1 in cells leads to the retardation of DNA replication and accumulation of unrepaired DNA intermediates, resulting in DNA double strand breaks (DSBs) and apoptosis. Therefore, targeting FEN1 could serve as a potent strategy for cancer therapy. In this study, we demonstrated that FEN1 is overexpressed in breast cancers and is essential for rapid proliferation of cancer cells. We showed that manipulating FEN1 levels in cells alters the response of cancer cells to chemotherapeutic drugs. Furthermore, we identified a small molecular compound, SC13 that specifically inhibits FEN1 activity, thereby interfering with DNA replication and repair in vitro and in cells. SC13 suppresses cancer cell proliferation and induces chromosome instability and cytotoxicity in cells. Importantly, SC13 sensitizes cancer cells to DNA damage-inducing therapeutic modalities and impedes cancer progression in a mouse model. These findings could establish a paradigm for the treatment of breast cancer and other cancers as well.
Keywords: Flap endonuclease 1 (FEN1), DNA replication and repair, Breast cancer, Chemotherapeutic drug, Small molecular inhibitor
Highlights
-
•
FEN1 is overexpressed in cancer cells and essential for cancer cell growth;
-
•
Down regulation of FEN1 leads to retarded cell growth and sensitizes cancer cells to chemotherapeutic agents;
-
•
SC13, a FEN1 specific inhibitor, inhibits cancer growth in vitro and in xenograft tumor mice.
Most anticancer agents used in clinic today kill cells by interfering DNA replication or inducing DNA damage, which in turn lead to cell apoptosis. However, cancer cells have evolved a compilation of highly effective DNA replication and repair systems to meet up the requirement of rapidly dividing of cancer cells and protect DNA against both endogenous and exogenous DNA damage. FEN1 has been shown to be an important factor in both DNA replication and repair pathways, making FEN1 a logical target for developing anticancer drugs as stand-alone agents for treating cancers that rely on its activity and as a therapy in combination with chemotherapeutic agents that cause DNA damage.
1. Introduction
DNA flap endonuclease 1 (FEN1) represents a unique class of structure-specific 5′ endonucleases (Shen et al., 2005). Unlike many other endonucleases that recognize a specific DNA sequence, FEN1 recognizes a specific DNA structure, independent of the DNA sequence (Shen et al., 2005). FEN1 has three distinct activities: 5′ flap endonuclease (FEN), exonuclease (EXO), and gap-dependent endonuclease (GEN) activities. FEN activity, the most dominant activity of FEN1, plays a major role in RNA primer removal during Okazaki fragment maturation of the lagging strand during DNA replication and in the removal of flap structures formed during long-patch (LP) base excision repair (BER) (Balakrishnan and Bambara, 2013). Thus, FEN1 has dual functions in DNA replication and repair and is essential for genome stability and integrity. Deficiencies in FEN1 function or deletion of the FEN1 gene have profound biological effects. Mouse model studies showed that FEN1 deficiency causes defects in DNA replication, failure of cell proliferation, and embryonic lethality (Kucherlapati et al., 2002, Zheng et al., 2007a). Moreover, mouse embryonic fibroblasts (MEFs) having defective FEN1 are sensitive to DNA damaging agents such as methyl methane sulfonate (MMS) and γ-radiation (Larsen et al., 2003, Zheng et al., 2007b).
Due to its fundamental role in DNA replication, FEN1 is required to support hyper-proliferation of cancer cells. Indeed, there is growing evidence that FEN1 expression is associated with the onset and progression of cancer. FEN1 is expressed at low levels in quiescent cells (Kim et al., 2000), but is highly expressed in proliferative tissues and cancers including lung (Nikolova et al., 2009), breast (Singh et al., 2008), gastric (Wang et al., 2014), prostate (Lam et al., 2006), pancreatic (Iacobuzio-Donahue et al., 2003) and brain cancers (Krause et al., 2005). Moreover, the level of FEN1 expression in cancer tissues has been correlated with increased cancer grade and aggressiveness (Abdel-Fatah et al., 2014). Thus, we propose that inhibiting FEN1 activity could suppress cancer cell growth.
Most chemotherapeutic drugs used clinically evoke cell apoptosis by inducing DNA damage. However, the high efficiency of DNA repair due to the overexpression of DNA repair proteins in cancer cells reduces the drug efficacy significantly (Fink et al., 1996, Fink et al., 1998). For example, the expression level of DNA polymerase beta (Pol β) has been correlated with resistance of cancer cells to chemotherapeutic drugs (Lawson et al., 2011). Cells with higher levels of DNA ligase IV exhibit reduced levels of γ-H2AX foci (an early marker of DNA damage in cells) upon treatment with DNA damage agents (Srivastava et al., 2012). Moreover, patients with DNA repair efficiency defects are more sensitive to chemotherapy (Riballo et al., 1999). Based on the roles of FEN1 in DNA repair, we speculate that inhibition of FEN1 could lead to the generation of DNA lesions, thus sensitize cancer cells to chemotherapy.
Breast cancer remains the most common cancer in females, and its incidence continues to rise (Hutchinson, 2010). There is an urgent demand for novel drugs effective in treating breast cancer. In this study, we showed that FEN1 is overexpressed in breast cancer. Using the MCF7 breast cancer cell line as a research model, we demonstrated that FEN1 is essential for proliferation and drug resistance in breast cancer cells. Furthermore, we identified a FEN1 inhibitor, SC13. SC13 blocks FEN1 activity specifically and impairs DNA replication and repair in vitro and in cells. SC13 suppresses cell growth, resulting in the accumulation of DNA double strand breaks (DSBs) in cells, thereby culminating into cytotoxicity. Finally, using mouse cancer models, we showed that SC13 impedes progression of cancer growth, causing a significant increase in the sensitivity of cancers toward chemotherapy.
2. Materials and Methods
2.1. Cell Lines and Cell Culture
All cell lines used in this study were from the American Type Tissue Collection and were cultured under conditions as directed by the product instructions.
2.2. Immunochemistry Analysis
Tissues were fixed in 10% formalin. Paraffin-embedded sections from tissue specimens were de-paraffinized and heated at 97 °C in 10 mM citrate buffer (pH 6.0) for 20 min for antigen retrieval. Primary antibodies used in immunocytochemistry were raised against FEN1. Immunoreactivities were analyzed by estimating the percentage of cells showing characteristic staining and the intensity of staining (Elakoum et al., 2014). The intensity of staining was graded as 1 (weak), 2 (medium), or 3 (strong). Results were scored by multiplying the percentage of positive cells (P) by the intensity (I) to obtain the Q-score (Q), which ranged between 0 and 300. A Q-score of 300 represented 100% of cells strongly stained (Q = P × I; maximum = 300) (Elakoum et al., 2014, Wu et al., 2011).
2.3. FEN1 Nuclease Activity Assay
The cleavage of DNA substrates by FEN1 was determined under the same conditions as those previously published (Guo et al., 2008, Sun et al., 2016). Double flap DNA substrate (a long 5′ flap and a short 3′ falp) has been reported as the best substrate for Flap activity (Finger et al., 2009, Tsutakawa et al., 2011, Kao et al., 2002, Friedrich-Heineken and Hubscher, 2004). Briefly, 32P-labeled DNA substrates were incubated with purified FEN1 in a buffer solution containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 5 mM MgCl2. The reactions were carried out at 37 °C for 30 min and were terminated with stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). The product and substrate were then separated by 15% PAGE and visualized by autoradiography.
2.4. DNA Replication Efficiency Assay
DNA replication efficiency of cells was measured by 3H incorporation assay (Sun et al., 2016). Briefly, 5 × 104 cells were seeded onto a 6-cm dish in Dulbecco's Modified Eagle Medium (DMEM) for 12 h. 3H-thymidine was added to a final concentration of 1 μCi/mL. Cells were incubated in 3H-thymidine-containing DMEM for a specific time period and washed with ice-cold PBS buffer. DNA was precipitated by treating cells with 10% ice-cold trichloroacetic acid and 10 mM thymidine (15 min, 4 °C). After washing extensively with PBS buffer, DNA was solubilized in 0.5 M NaOH. Radioactivity levels in the sample were measured using a liquid scintillation counter.
2.5. Okazaki Fragment Maturation Assay
Okazaki fragment maturation was assayed according to published protocols (Zheng et al., 2007a, Zheng et al., 2007b, Jaiswal et al., 2011). Briefly, cell extracts were incubated with specific gapped substrates in a reaction buffer containing 5 μCi [α-32P]dCTP and 50 μM each of dATP, dGTP, dTTP, and 2 mM ATP. Reactions were carried out (60 min, 37 °C) and products were separated by 15% denaturing PAGE, followed by autoradiography.
2.6. Base Excision Repair Assay
The BER assay was performed as described previously (Guo et al., 2009, Zhou et al., 2016). Reactions were carried out in 20 μl of reaction buffer B (40 mM HEPES-KOH [pH 7.8], 70 mM KCI, 7 mM MgCl2, 1 mM dithiothreitol, 0.5 mM EDTA, 2 mM ATP, 50 μM each of dATP, dTTP, and dGTP, and 2 μCi [α-32P]-dCTP). For SP reconstitution with purified proteins, Uracil-DNA Glycosylase (UDG, 8 ng), APE1 (2 ng), Ligase IIIα (20 ng) and various amounts of Pol β (0–5 ng), were mixed and incubated with the SP-BER substrate Pol β-U (Fig. 4e). For LP-BER, the Pol β-F substrate (Fig. 4f) was incubated with a mixture of APE1 (2 ng), Pol β (0–5 ng), FEN1 (2 ng), and Ligase I (20 ng). For cell extract reconstitution (Fig. 4c), the LP-BER DNA substrate was incubated with the whole cell extract (WCE) (5 μg). Reactions (30 min, 37 °C) were then stopped by adding an equal volume of the gel loading buffer and visualized by autoradiography.
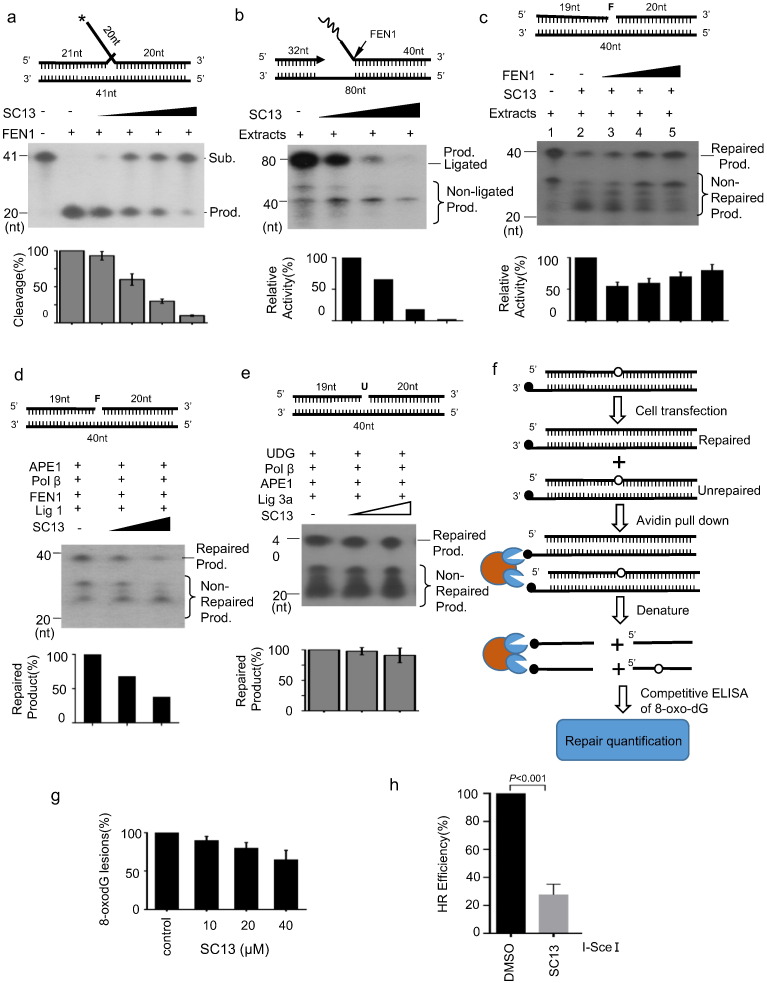
Fig. 4.
Effects of SC13 on DNA repair efficiency. a. Effects of SC13 on FEN1 activity. b. Okazaki fragment maturation assay. A gap substrate with a RNA-DNA flap was incubated with cell lysates and increasing amounts of SC13. Reconstitution of long-patch (LP) base excision repair (BER) with cell lysate (c) or purified proteins (d). A tetrahydrofuran (THF)-containing oligo was used to mimic the LP-BER substrate. e. Reconstitution of short-patch BER with purified proteins. An uracil-containing oligo was used to mimic the short-patch BER substrate. The top of each panel shows the schematic structures of the corresponding DNA substrates. The middle panel shows PAGE-separated products, and the bottom panel shows the relative percentage of cleavage of substrate. The values represent the mean ± SD of three independent assays. f. Procedure of LP-BER assay in cells. A DNA oligo containing the damaged DNA lesion 8-oxo-dG was transfected into MCF7 cells; 4 h later, cells were lysed, and released 8-oxo-dG was determined by ELISA. g. Effects of SC13 on LP-BER in cells. h. Cell-based homologous recombination (HR) repair assay. U2OS-DR-GFP cells were transfected with I-Sce I plasmid and treated with SC13 or DMSO (as a negative control).
2.7. In vivo 8-Oxo-dG BER Assay
An extrachromosomal assay system (Sundaresakumar, 2009) using a biotin-tagged oligonucleotide DNA substrate containing an 8-oxo-dG lesion that can be repaired by LP-BER (Sattler et al., 2003). The 8-oxo-dG BER assay uses a double-stranded oligonucleotide DNA substrate that consists of a biotinylated DNA strand that is base paired to an untagged DNA strand containing a single 8-oxo-dG lesion. The substrate DNA is transfected into MCF7 cells and incubated for 4 h to allow repair of the 8-oxo-dG lesion. SC13 was added to the culture medium to test its effects on 8-oxo-dG repair. The cells are then lysed and the biotinylated substrate DNA is captured using streptavidin-coated magnetic beads. In the presence of a magnet, the strand containing the 8-oxo-dG lesion is denatured and isolated from the streptavidin-bound biotinylated strand. The isolated DNA is then quantified and subjected to an 8-oxo-dG competitive ELISA using a competitive 8-oxo-dG ELISA kit (Trevigen, Cat. No.4380-192-K).
2.8. HR Reporter Assays
U2OS cells with DR-GFP integration was used to measure homologous recombination efficiency as described. Specifically, 3.5 × 105 U2OS-DR-GFP cells were seeded in six-well plates per well and 24 h later transfect plasmid pCBASce (an I-SceI expression vector) with Lipofectamine 3000 under manufacture's protocol. Culture medium contained 2 μM SC13, which will not cause significant cell toxicity. 48 h after transfection, cells were harvested and subjected to flow cytometry analysis to assess the GFP positivity. Relative HR efficiency was calculated and compared using CFCS and GraphPad PRISM software.
2.9. Drug Sensitivity Assay
The sensitivity to DNA damage-inducing reagents was determined by cell growth inhibition assays (Guo et al., 2009). MCF7 cells were seeded (1500/well), incubated (overnight, 37 °C), treated (1 h, 37 °C) with multiple dilutions of H2O2, washed in fresh medium (DMEM containing 10% fetal bovine serum) and incubated (72 h) under normal growth conditions (37 °C, 5% CO2). The number of viable cells was determined by the CellTiter 96 AQueous one-solution cell proliferation assay (Promega). At least four replications were performed for each clone, and the means were calculated. Data were expressed as the percentage of growth relative to untreated controls.
2.10. Cellular Transformation Assay
The focus formation assay was conducted according to a previous report (Chung et al., 2015). The presence of foci was visually evaluated after staining the cells with 0.05% crystal violet. The anchorage-independent growth assay in soft-agar was described previously (Bose et al., 2013). Briefly, the cells (3000 cells per 35 mm well) were suspended in complete medium containing 0.35% agarose. The cells were grown on tissue culture dishes containing a 2 mL layer of solidified 0.7% agar in a complete medium. After 10 days, the number of colonies was quantified from micrographs taken at random locations on the plate (original magnification, × 20).
2.11. Xenograft Studies
Female homozygous, 6-week-old, nude mice were used in this study. MCF7 cells were harvested, and single-cell suspensions with > 95% viability (5 × 106 cells) diluted with equal volumes of Matrigel (BD Biosciences) were injected subcutaneously into the right flank of each mouse. After the cancers were established, drugs were administered intraperitoneally daily for five consecutive days. Cancer volume was measured weekly in each group. Mice were euthanized when the cancer volume reached ~ 1000 mm3. The mice were housed and maintained under standard NIH protocol.
3. Results
3.1. FEN1 Is Overexpressed in Breast Cancer and Co-Related to Malignancy
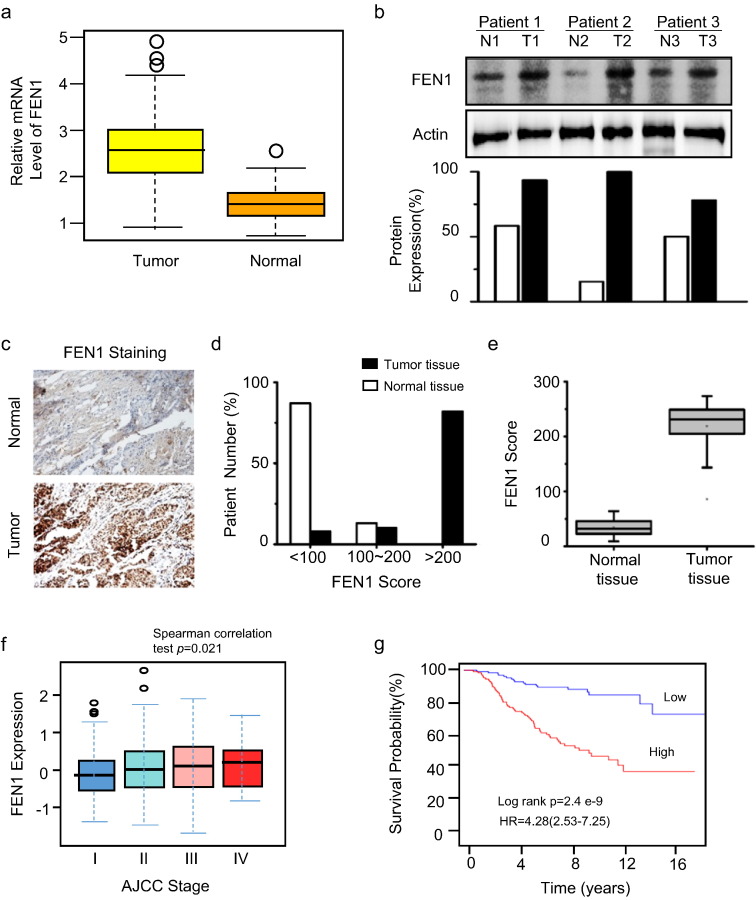
According to the Cancer Genome Atlas (TCGA) database, FEN1 is overexpressed in various types of cancers, including breast, prostate, gastric, lung, and colorectal cancers (Figs. 1a and S1a), which is consistent with our previous report (Singh et al., 2008). We chose breast cancer for further study, as it is the most common cause of death from cancer in females (Matsen and Neumayer, 2013). To verify the data from the TCGA database, we collected breast cancer samples from surgical excisions and compared the levels of FEN1 protein in cancer and normal tissues from the same patient using western blotting (Fig. 1b). We found FEN1 was highly expressed in breast cancer tissue compared to the corresponding normal tissue (Fig. 1b). In addition, FEN1 displayed significantly stronger staining in cancer specimens than in their healthy counterparts in immunohistochemistry assays (Fig. 1c). The intensity of staining was graded 1 for weak, 2 for medium and 3 for strong. Results were scored by multiplying the percentage of positive cells (P) by the intensity (I) that is by using the quick score (Q). A Q-score between 0 and 300 is obtained where 300 is equal to 100% of cells strongly stained (Q = P × I, maximum = 300). Score of < 100 is considered weak, and > 200 is considered strong. While82% of patients had a FEN1 score < 100 (95% CI, 78–86%) in normal tissue, a significantly lower proportion of patients (8%) displayed a FEN1 score < 100 in tumor tissues (95% CI, 3–13%; p < 0.001). Conversely, the proportion of patients with FEN1 scores > 200 was significantly higher in tumor than normal tissue (78% vs. 0%; p < 0.001; Fig. 1d). Moreover, the median FEN1 expression was elevated significantly in cancer tissues compared to that in the corresponding normal tissues (p < 0.001; Fig. 1e). Furthermore, the FEN1 protein level was overexpressed in various breast cancer cell lines such as MCF7, MDA-MB-231 and MDA-MB-435, when compared to the normal breast epithelial cell line, MCF10A (Fig. S1b).
Fig. 1.
DNA flap endonuclease 1 (FEN1) overexpression in breast cancer. a. Statistical analysis of FEN1 mRNA expression levels in breast cancer from the TCGA database. b. Western blot to determine FEN1 protein levels in normal and breast cancer tissues. N, normal; T, tumor. Specimens of surgically removed tumors were assayed for FEN1 protein expression. The bottom panel shows the quantification. c. Immunohistochemical staining of FEN1. d. Number of patient samples with FEN1 scores > 200 is significantly higher in tumors than in control tissue. e. Median FEN1 scores are elevated in tumor tissue. f. Correlation of FEN1 expression and breast cancer stages. g. Associations between FEN1 expression and patient prognostics.
Next, we investigated whether the FEN1 expression level is correlated to the malignancy of breast cancers. Data from the ATCG database indicated that the malignancy grade rose with increasing FEN1 expression levels in breast cancers (Fig. 1f), suggesting that FEN1 overexpression is associated with high levels of malignancy.
To validate this conclusion, we employed a tissue array of 48 breast cancer tissues with various malignancy grades (Table S1). Table S1presents the expression pattern of FEN1 in various breast cancer tissues. The results showed that high expression of FEN1 is associated with a statistically significant probability of increased malignancy (p < 0.01). The statistical analysis of the FEN1 status in relation to TNM stage and tumor grade was performed using the parametric Fisher's exact test (Table S1). High expression of FEN1 appeared to associate in a statistically significant manner with highly malignant tumors (p < 0.05) and tumors of advanced TNM stage (p < 0.05). Moreover, consistent with this result and our previous report (Wang et al., 2015), Kaplan-Meier analyses revealed that breast cancer patients with high expression of FEN1 have poor prognoses (Fig. 1g). These results suggest that FEN1 is associated with onset and progression of breast cancer.
3.2. Overexpression of FEN1 Promotes Cell Growth and Induces Foci Formation
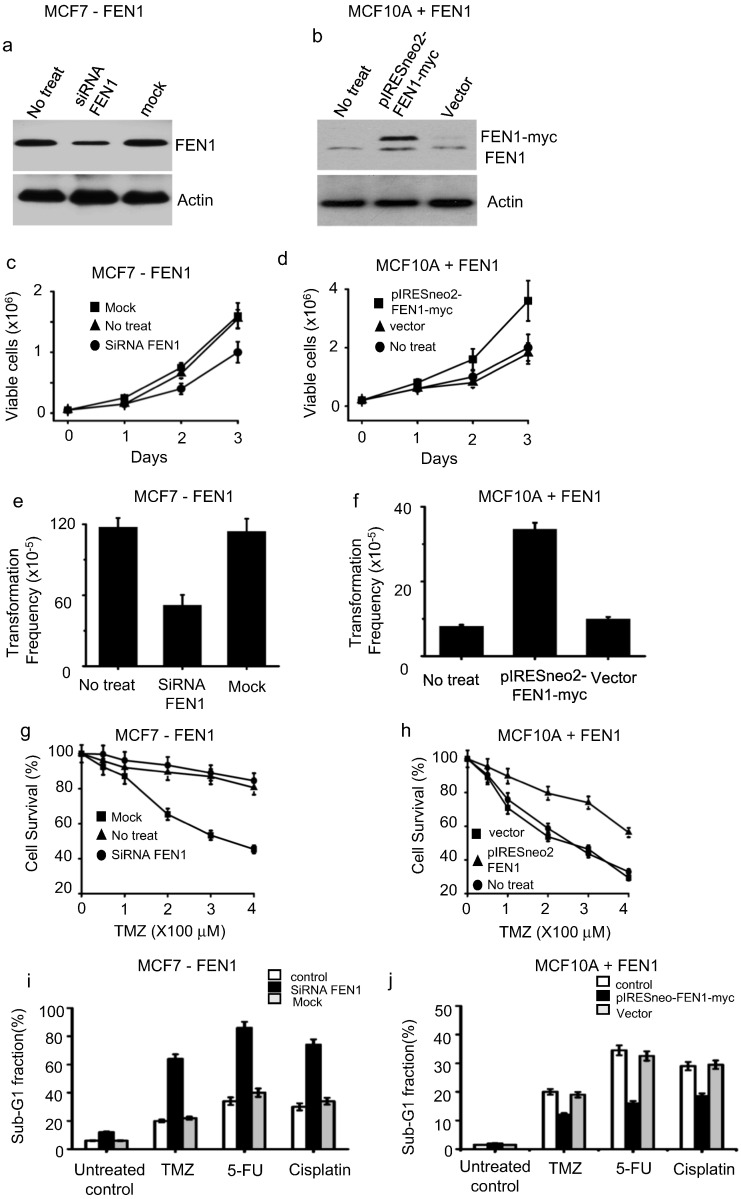
To test whether FEN1 is essential for rapid cell growth and promotes foci formation, FEN1 is suppressed in MCF7 (high endogenous FEN1 expression, Figs. S1b, 2a) or overexpressed in MCF10A (low endogenous FEN1 expression, Figs. S1b, 2b). Suppressing FEN1 expression in MCF7 led to retarded cell growth (Fig. 2c) and foci formation (Fig. 2e) while overexpression of FEN1 in MCF10A promoted cell growth (Fig. 2d) and induced foci formation (Fig. 2f). This observation was confirmed in other breast cancer cell lines (T47D, MDA-MB-231, and HCC1937) and cell lines from various cancer types such as lung (A549) and colorectal (HCT8) cancer (data not shown), indicating that this phenomenon is not cell-line-specific. These data suggest that FEN1 is essential for the growth of cancer cells.
Fig. 2.
FEN1 is essential for cell proliferation and contributes to drug resistance in cancer cells. FEN1 was (a) knocked-down in MCF7 or (b) overexpressed in MCF10A cells. The expression level of FEN1 was determined by western blot analysis using anti-FEN1 antibody. Cell growth rate assay of cells in which FEN1 was (c) suppressed or (d) overexpressed. Foci formation assay in cells with (e) FEN1 suppression or (f) overexpression. g. FEN1 knockdown sensitizes MCF7 cells to temozolomide (TMZ). h. FEN1overexpression inMCF10A cells caused increased resistance to TMZ. Cells with FEN1downregulation (i) or FEN1 overexpression (j) were harvested and stained, and the percentage of sub-G1 fraction was determined by flow cytometry.
3.3. FEN1 Regulates the Sensitivity of Cancer Cells to DNA Damage-inducing Agents
As FEN1 is involved in DNA repair, we speculated that manipulation of FEN1 could alter the response of cancer cells to chemotherapeutic drugs that induce DNA lesions. To test this, FEN1 level was manipulated in cells and the survival rate of resulted cells were determined after exposed to temozolomide (TMZ), a commonly used DNA damage-inducing cancer drug (Chamberlain, 2010). We found that FEN1-knockdown-treated cells show greater sensitivity to TMZ (Fig. 2g), whereas FEN1-overexpressing cells demonstrated higher resistance to this drug (Fig. 2h) when compared to their own control cells. To further test whether FEN1 expression levels could impact the response of cancer cells to other DNA damage-inducing agents, we determined the sub-G1 ratio (apoptotic fraction of cells) of MCF7 after treatment with TMZ, fluorouracil (5FU) and cisplatin. We found that FEN1 knockdown augmented cell apoptosis induced by TMZ, 5FU, and cisplatin (Fig. 2i). Overexpression of FEN1 prevented apoptosis induced by these drugs (Fig. 2j). These data indicated that alterations in FEN1 levels might impact the tumor cell response to anticancer drugs.
3.4. Screening of Small Molecular Compounds to Inhibit FEN1
Based on the observations above, FEN1 is essential for cancer cell growth and determines the drug response of cancer cells. It is rational to speculate that inhibiting FEN1 function could be an effective strategy for cancer therapy. Therefore, FEN1 inhibitors could serve as potent anticancer drugs, which could be used alone to suppress cancer cell growth or combined with DNA damage-inducing agents to augment cell apoptosis.
A previous study by Tumey et al. showed that N-hydroxyl urea derivatives possess inhibitory action against FEN1 (Tumey et al., 2005). Compound #20, with an IC50 of 3 nM to FEN activity, is the most potent FEN1 inhibitor tested in vitro. However, its ability to suppress the growth of cancer cells is limited when used alone (Tumey et al., 2005). To generate a more potent anti-FEN1 compound, we designed two series of six novel small molecules based on the N-hydroxyl urea derivative #20 (Figs. 3a and S2a). The pharmacophores of the two series were pyrrolo[3.2-d] pyrimidine (SC11, SC12, and SC13) and thieno[3.2-d] pyrimidine (SC21, SC22, and SC23), respectively. To investigate the structure-activity relationship (SAR) and to improve the cell permeability of target compounds, the hydrophilic hydroxyl groups of urea were substituted by hydrophobic phenyl, benzyl, and n-propyl building blocks (Figs. 3a and S2a). Among six small molecules, SC13 (shown in Figs. 3a and 4a) displayed comparable inhibitory effects on FEN1 activity to the N-hydroxyl urea derivative #20 (Supplementary Fig. S3) but improved cell permeability.
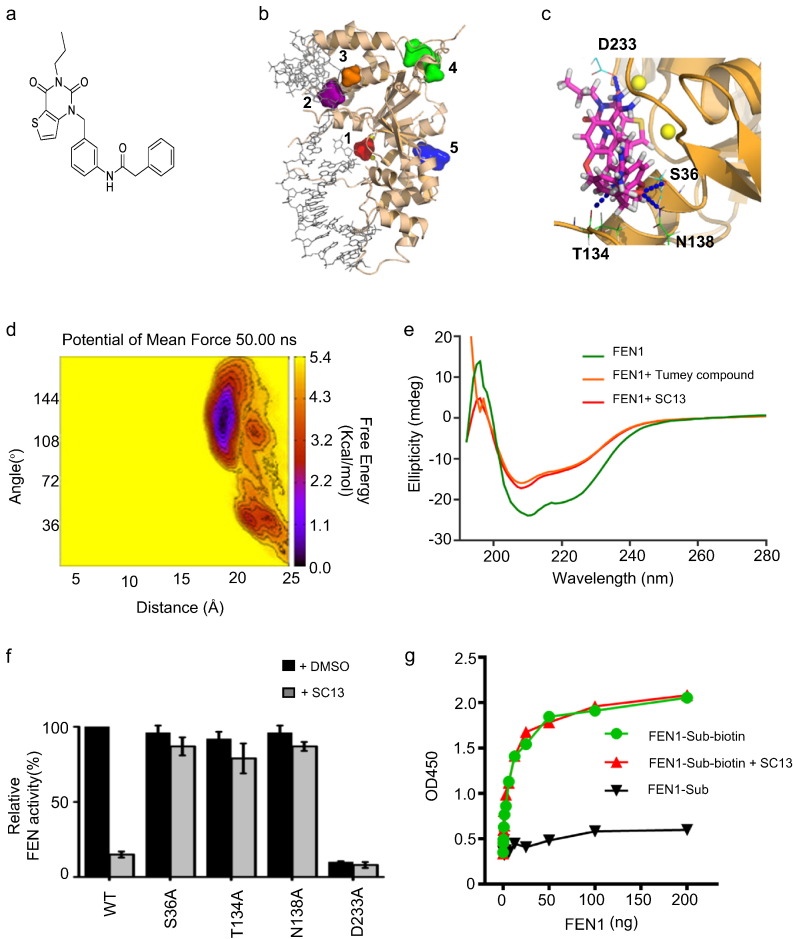
Fig. 3.
Design and characterization of SC13. a. Chemical structure of SC13. b. Best druggable sites of FEN1 predicted by Druggable Site Prediction (DSP). c. Detailed docking mode of SC13 bound to FEN1. Four hydrogen bonds (blue dots) are formed between SC13 compound and S36, T134, N138, and D233 residues on the FEN1 protein. d. Potential of mean force of SC13 binding with FEN1 protein predicted by implementing 50 ns meta-dynamics simulation. e. Circular dichroism spectroscopy analysis of FEN1/SC13 binding. f. Inhibitory effects of SC13 on FEN1 mutations. g. Effects of SC13 on DNA substrate binding.
To understand how SC13 interferes with FEN1, we performed docking studies to elucidate the interacting site of SC13 to FEN1. Five best druggable sites of the FEN1 protein were predicted using our in-house developed Druggable Site Prediction (DSP) algorithm (Gu et al., 2015) (Fig. 3b). Approximately 65% of the FDA-approved drugs showed a preference for binding the best druggable site (site 1, shown in red, Fig. 3b), while 3–7% of the drug molecules showed a binding preference for the other four druggable sites. SC13 binds FEN1 protein at the best druggable site by forming four hydrogen bonds with S36, T134, N138, and D233 (Fig. 3c). To confirm the binding pocket of SC13, meta-dynamics simulation was carried out (Laio and Parrinello, 2002) (Fig. 3d). The 50-ns molecular dynamics simulation shows that SC13 remains in the pocket with a binding affinity of approximately − 4.2 kcal/mol.
To confirm the binding of SC13 to FEN1, we performed circular dichroism spectroscopy analysis. The results showed a clear shift in the spectrum upon addition of SC13 to FEN1, compared to the control (Fig. 3e), suggesting a physical interaction of SC13 with FEN1. The data also showed that Tumey #20 and SC13 bound to FEN1 and resulted in a similar conformation change (Fig. 3e).
To further validate that S36, T134, N138, and D233 residues are involved in the SC13/FEN1 interaction, we generated the following FEN1 mutations: S36A, T134A, N138A, and D233A. The results showed that while the FEN activity of these mutants remained intact (with the exception of D233A, which was inactive), the inhibitory effects of SC13 were disrupted (Fig. 3f), suggesting that these residues are involved in SC13-related FEN1 inhibition activity.
For example, as displayed in Fig. 3f, the S36A mutation itself does not affect FEN1 activity significantly (slightly drops to about 95%). Meanwhile, as the binding model shown in Fig. 3c, S36 may form hydrogen bond with SC13 molecule to enhance the inhibitor binding. Thus, S36 is a good residue for mutagenesis to validate the model of FEN1 in complex with SC13: its mutation would not affect FEN1 activity while the mutation might interrupt the binding of SC13 inhibitor. Indeed, we see that the S36A mutated FEN1 activity does not drop significantly when SC13 is presence (slightly drops to about 90%), while in wild-type the FEN1 activity drops dramatically to about 15%. This implies that the S36A mutation effectively blocks SC13 bound to FEN1 protein. Consequently, the S36A mutated FEN1 activity should not be affected significantly by SC13 anymore. Therefore, S36 should be one of the key residues to directly contact with the FEN1 inhibitor.
To test whether SC13 binding to the active center of FEN1 impaired DNA substrate binding, we performed a DNA-FEN1 binding assay. Surprisingly, although SC13 binds to the FEN1 active center, it does not interfere with FEN1-substrate binding (Fig. 3g). Moreover, we found that SC13 did not exert any inhibitory effects on EXO activity or GEN activity at the IC50 of FEN activity (Fig. S3c,d). Therefore, SC13 inhibit FEN activity of FEN1 specifically.
To determine whether SC13 is a specific inhibitor of FEN1 activity, we investigated whether it also blocks the activity of other BER enzymes such as apurinic/apyrimidinic endonuclease 1 (APE1), Pol β, and DNA ligase I. We also tested the effects of SC13 on DNase I. The results showed that SC13 did not inhibit the activities of these enzymes (Figs. S2b–d, S3d), indicating that the inhibitory effect of SC13 was highly specific for FEN1.
3.5. SC13 Interferes with Okazaki Fragment Maturation and Long Patch Base Excision Repair
As described previously, FEN1 is involved in the maturation of Okazaki fragments and long patch base excision repair (LP-BER). To determine whether SC13 affects Okazaki fragment maturation, SC13 was added to the in vitro reaction system. The results showed that SC13 decreased the efficiency of RNA primer flap removal and prevented DNA ligation (Fig. 4b), indicating the Okazaki fragment maturation efficiency was impaired by SC13.
To test whether SC13 affected the overall LP-BER efficiency, we performed an in vitro LP-BER reconstitution assay using cell extracts (Fig. 4c) or purified proteins (Fig. 4d). We found that while cell extracts repaired DNA damage efficiently, as demonstrated by an intense 40-nt band (Fig. 4c, lane 1), SC13 reduced the amount of repair dramatically (Fig. 4c, lane 1 vs. 2). Interestingly, the addition of purified FEN1 restored the LB-BER activity (Fig. 4c, lanes 3–5). These results indicated that the inhibitory effect of SC13 on LP-BER is FEN1 dependent. Moreover, SC13 inhibited LP-BER (Fig. 4d), but not short patch BER (Fig. 4e) in a reconstitution assay with purified protein, further supporting that SC13 interferes with FEN1 activity specifically (FEN1 is not required in the short patch BER assay).
To further evaluate the effects of SC13 on BER inside the cells, we adopted an extrachromosomal assay system (Sundaresakumar, 2009) using a biotin-tagged oligonucleotide DNA substrate containing an 8-oxo-dG lesion that can be repaired by LP-BER (Sattler et al., 2003). The substrate DNA is transfected into target cells and incubated (4 h) to allow repair of the lesions. The DNA is then captured following cell lysis and the disappearance of 8-oxo-dG lesions is monitored using competitive 8-oxo-dG ELISA (Fig. 4f for assay procedure). SC13 was added to the cell culture medium at the incubation step. To exclude the general toxicity of SC13 on cells, we tested the effects of SC13 on cell proliferation at the same condition. We found no cytotoxicity was observed among the cells that were treated with SC13 for 4 h up to 250 uM (Fig. S4). In this manuscript, we chose up to 50 uM SC13 for 8-oxo-dG repair inhibition assay and all the other cell-based assays in this study.
As shown in Fig. 4g, upon addition of SC13, a decrease in 8-oxo dG repair was observed, suggesting that SC13 inhibited endogenous BER at the intracellular level (Fig. 4g). As FEN1 was recently reported to be required for homologous recombination repair (Fehrmann et al., 2015), we speculated that SC13 might also interfere in HR repair. Indeed, in the assay with a stably integrated GFP-based homologous recombination substrate (Gunn and Stark, 2012), SC13 clearly impaired HR-mediated repair (Fig. 4h).
3.6. SC13 Retards DNA Replication and Suppresses Cell Growth
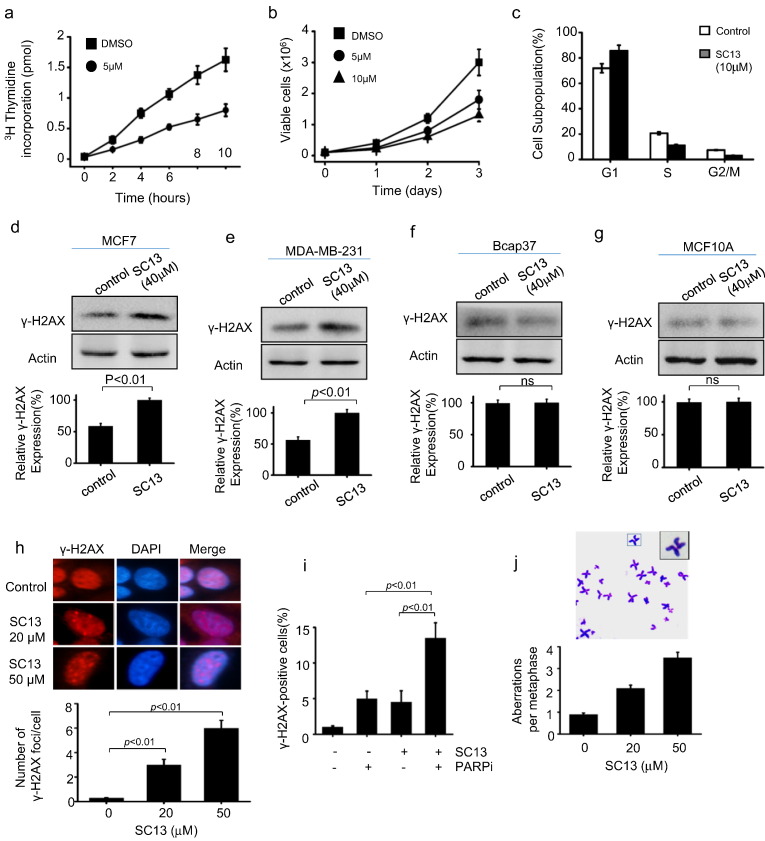
The SC13-induced retarded efficiency of Okazaki fragment maturation (Fig. 4b) might affect DNA replication in cells. Therefore, we evaluated the DNA replication rate of cells with or without SC13 treatment using a 3H-thymidine incorporation assay. The data showed that SC13 caused a significant reduction in 3H-thymidine incorporation, indicating DNA replication was inhibited by SC13 (Fig. 5a). As a consequence of this reduction in DNA replication, the overall cell proliferation rate was reduced (Fig. 5b).
Fig. 5.
Inhibition of FEN1 retarded DNA replication and led to the accumulation of DNA damage. a. Thymidine incorporation assay. b. Cell growth assay. c. FACS to determine the effects of SC13 on cell cycle profile. Cell staining by γ-H2AX. Western blotting assay to determine the γ-H2AX level before and after SC13 treatment in (d) MCF7, (e) MDA-MB-231, (f) Bcap37 and (g) MCF10A cells. h. Cell staining to determine γ-H2AX foci in MCF7 cells treated by SC13 (20 μM and 50 uM), or (i) with co-treatment by SC13 (10 μM) and PARP inhibitor (10 μM). j. Effect of SC13-induced FEN1 inhibition on the induction of genome instability. Giemsa-stained metaphase spreads showing the chromosomal breaks generated upon SC13 treatment.
A defect in the DNA replication process might arrest cells in G1 phase and decrease S and G2/M phases (Sclafani and Holzen, 2007). To test the effects of SC13 on the cell cycle profile, we carried out a FACS assay. As shown in Fig. 5c, SC13 treatment caused an accumulation of cells in G1 phase but a decrease of cells in S and G2/M phases compared to the control (Fig. 5c). These data suggest that SC13 impeded cancer cell growth, consistent with an impaired Okazaki fragment maturation and delayed cell cycle progression.
3.7. SC13 Leads to Generation of Unrepaired DSBs and Chromosome Breaks in Cells
Previous reports indicated that defects in the Okazaki fragment maturation process during DNA replication or ligation during DNA repair could lead to the accumulation of DNA DSBs (Pascucci et al., 2005). Therefore, SC13-treated cells would be expected to display a higher level of DNA damage.
To test this, we treated FEN1 proficient cells MCF7 with SC13, followed by immunofluorescence and western blotting using anti-γ-H2AX. Results showed that in the presence of SC13, the levels of γ-H2AX in cells were higher than that in the control (Fig. 5d). Similarly, SC13-treatment also induce the accumulation of γ-H2AX in MDA-MB-231, also breast cancer cell line (Figs. 5e, S1). These data indicated that SC13 treatment resulted in accumulation of DNA DSBs in cells. However, when SC13 were applied to cells with low FEN1 expression, including Bcap37 (Fig. 5f) and MCF10A (Fig. 5g), no significant change was observed after SC13 treatment, suggesting that SC13-induced DSBs is FEN1-dependent manner. Consistently, cells with FEN1-knockdown do not response to SC13 treatment (Fig. S5).
HR repair is the major pathway to repair DSBs in cells (Dudas and Chovanec, 2004). Therefore, we speculated that inhibition of HR could augment the effects of SC13 to generate unrepaired damage in cells. Indeed, we observed increased numbers of DNA breaks when the FEN1 inhibitor was combined with PARP1 inhibitor (Fig. 5i). Moreover, the accumulation of DNA strand breaks in the genome may cause chromosome breaks (van Gent et al., 2001). Therefore, we determined whether SC13 could induce such chromosomal aberrations by counting the number of chromosomes in SC13-treated MCF7 cells. We found that the SC13-treated cells were more susceptible to the generation of chromosomal breaks (Fig. 5j). Thus, SC13 interferes with DNA replication and repair in cells, leading to the accumulation of unrepaired DSBs and chromosome breakage.
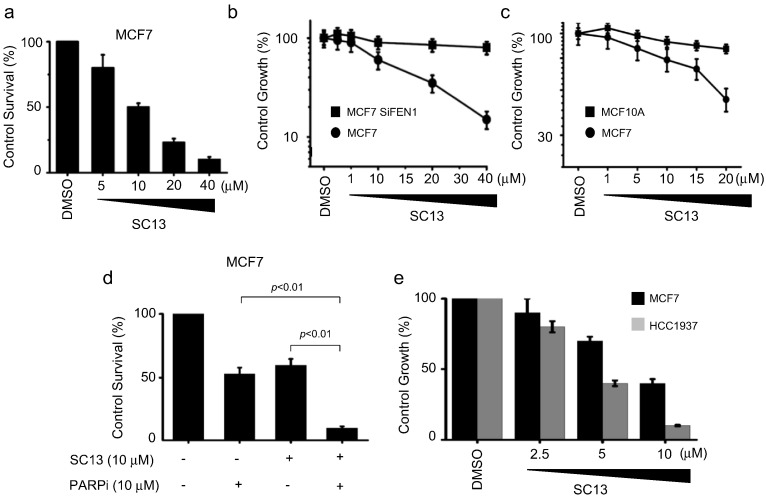
3.8. SC13 Induces Cytotoxicity by Targeting FEN1
The accumulation of unrepaired DSBs and chromosomal breaks induce cell apoptosis (Roos and Kaina, 2013). To evaluate whether SC13 treatment leads to cell death, we treated MCF7 cells with increasing concentrations of SC13. We showed a dose-dependent decrease in cell proliferation of MCF7 cells, indicating the cytotoxicity of SC13 to cancer cells (Fig. 6a). However, SC13-mediated cytotoxicity was limited among cells in which FEN1was knocked down (Fig. 6b) or in which FEN1 expression level is low (MCF10A cells) (Figs. 6c, S1). These data indicated that SC13 induces cell death by targeting FEN1.
Fig. 6.
Cytotoxicity induced by SC13 is correlated with FEN1 expression level in cells. a. Cytotoxicity induced by SC13 on MCF7, as measured by MTT assay. b. Effect of SC13 on cell proliferation of MCF7 before or after FEN1 knockdown. c. Comparison of effect of SC13 on cell proliferation of MCF7 and MCF10A. d. Cytotoxicity assay of MCF7 cells co-treated with SC13 and PARP inhibitor. e. Effects of SC13 on MCF7 (WT) and HR-deficient cell line HCC1937 (BRCA−/−).
As SC13 induced more DNA breaks in cells treated with a PARP1 inhibitor (Fig. 5i), we speculated that SC13 is more cytotoxic to cells when HR is inhibited or defective. Indeed, consistent with Fig. 5i, the combination of SC13 and PARP1 inhibitors led to more cell death than SC13 or PARP1 inhibitor alone (Fig. 6d). HCC1937 is a breast cancer cell line harboring a homologous BRCA1 mutation (Tomlinson et al., 1998). It synthesizes a truncated BRCA1 protein which is the product of a disease-producing mutant allele (5382insC). The HCC1937 cell line is HR-deficient and we showed that this cell line is more sensitive to SC13 than MCF7 cells (BRAC1+/+). These data indicated that in the absence of efficient HR, DSBs that were accumulated due to blockage of FEN1 remained unrepaired and led to enhanced cell death (Fig. 6e). These results support previous reports, which showed a synthetic lethal interaction between FEN1 and HR pathway proteins (Mcmanus et al., 2009, Van Pel et al., 2013).
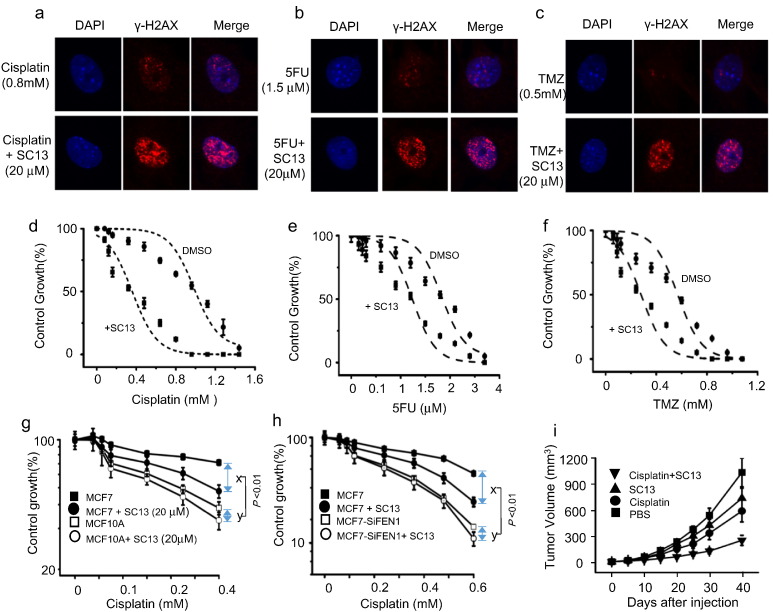
3.9. SC13 Enhances the Sensitivity of Cancer Cells to Chemotherapeutic Agents
Based on the data above, we speculated that SC13 treatment combined with chemotherapeutic drugs could enhance the frequency of DSBs in cancer cells. The results showed that chemotherapeutic drugs generated a higher number of γ-H2AX foci per cell upon addition of SC13, as compared to drug alone (Fig. 7a–c). We further speculated that combining the use of SC13 and chemotherapeutic drugs that induce DNA damage could enhance the sensitivity of cancer cells. To test this, we determined the cell sensitivity to SC13 alone or in combination with chemotherapeutic drugs. The results showed that the combinations of SC13 with cisplatin, TMZ, or 5FU increased the sensitivity of cancer cells to these drugs (Fig. 7d–f, respectively). However, the sensitivity to the combination treatment was reduced in cells where FEN1 is low expressed (Fig. 7g) or knockdown (Fig. 7h). These results demonstrated that SC13 in combination with additional chemotherapy drugs could be used as a more effective strategy for treating cancers.
Fig. 7.
SC13 enhances the sensitivity of cancer cells to chemotherapeutic agents. Cell staining with γ-H2AX antibody in MCF7 cells following treatment with (a) cisplatin, (b) 5FU, (c) TMZ alone, or with SC13. Drug sensitivity assay of MCF7 cells following treatment with (d )cisplatin, (e) 5FU, (f) TMZ alone, or with SC13. g. Comparison of sensitization of MCF7 and MCF10A to cisplatin by co-treatment with SC13. h. Effects of FEN1 knockdown on SC13 response of MCF7 cells. x, enhancement of growth inhibitory effect in MCF7 cells by co-treatment with SC13. y, enhancement of growth inhibitory effect in MCF7-SiFEN1 cells by co-treatment with SC13. i. SC13 combination with cisplatin decreases the growth of cancers in a xenograft model.
To verify the in vitro results that showed efficacy with the combined treatment of SC13 and DNA damage reagents, we performed an in vivo xenograft study using nude mice, as described in Fig. 7i. Ten days after cell injection, the drugs were given intra-peritoneally daily for five consecutive days, and the cancer growth inhibition was monitored up to 40 days. The tumor volume increased in the control group in a time-dependent manner (Fig. 7i), but decreased in mice treated with cisplatin or SC13 alone. However, the tumor inhibitory effect was much more pronounced when SC13 was combined with cisplatin, suggesting that the combination treatment of SC13 enhances the therapeutic efficacy of chemotherapy drugs in a xenograft cancer model.
4. Discussion
Chemotherapy remains the most powerful tool for cancer treatment (Nguyen et al., 2014). Cancer cells are characterized by their ability to divide rapidly. As a critical enzyme for DNA replication, overexpression of FEN1 is believed to be a biomarker of cancer cells (Wang et al., 2014, Abdel-Fatah et al., 2014). Indeed, we and other groups demonstrated that FEN1 is overexpressed in various types of cancers (Fig. 1). We hypothesized that inhibition of FEN1 could suppress cancer growth by blocking DNA synthesis. Supporting our hypothesis, siRNA-mediated downregulation of FEN1 in MCF7 cells retarded cell proliferation significantly (Fig. 2c). The FEN1 inhibitor SC13 also impaired DNA replication and cell growth, leading to the accumulation of cells in G1 phase (Fig. 5a–c).
Besides blocking cell division, most anticancer agents used clinically cause DNA lesions to induce cell apoptosis. However, in many cases, an elevated DNA repair capacity in cancer cells leads to drug resistance and severely limits the efficacy of chemotherapeutic drugs (Kaina and Christmann, 2002). Therapeutic efficacy is determined by the balance between DNA damage and repair. Given the roles of FEN1 in DNA repair, we speculated that manipulation of FEN1 in cancer cells could alter the response of cancer cells to DNA damage agents. Indeed, we demonstrated that cancer cells with high FEN1 levels are more resistant to DNA damage agents such as TMZ, cisplatin, and 5FU (Fig. 2i–j). At the same time, FEN1knockdown sensitizes cancer cells to DNA damage. Furthermore, applying the FEN1 inhibitor SC13 augmented the cytotoxicity of anticancer drugs, both in cells and in a xenograft cancer mouse model (Fig. 5, Fig. 6, Fig. 7).
The dual function of FEN1 makes it an ideal target for cancer therapy. The FEN1 inhibitor can be used independently to suppress DNA replication, inducing DNA damage and apoptosis of cancer cells. Moreover, FEN1 inhibitor can also be combined with other DNA damage-inducing agents. In addition to suppressing cell growth by inhibiting DNA replication, FEN1 inhibitor can also lead to the accumulation of DNA DSBs and genome instability (Fig. 5h, j). Thus, the cytotoxicity induced by FEN1 inhibitor could be due to the combination of defects in DNA replication and repair.
HR repair is responsible for repairing DNA DSBs in cells (Dudas and Chovanec, 2004). Therefore, the suppression of FEN1 was expected to selectively sensitize cancer cells that are defective in HR. Indeed, the BRAC−/− breast cancer cell line HCC1937 is more sensitive to the FEN1 inhibitor SC13 when compared to the BRAC+/+ breast cancer cell line MCF7. The combination of SC13 and PARP1 inhibitors led to accelerated cell apoptosis compared to that observed in cells treated with SC13 or PARP1 inhibitor alone (Figs. 5i, 6d). These data indicated that FEN1 is an effective target for developing synthetic lethal interactions involving the HR pathway.
Besides the genes involved in the HR pathway, > 250 genes are predicted to have a synthetic lethal interaction with FEN1(Van Pel et al., 2013, Mcmanus et al., 2009). FEN1 suppression demonstrated synthetic lethality with Rad54B in Rad54B-deficient colorectal cancer (CRC) cell lines, and selectively sensitized hTERT-deficient ALT (Alternative pathway for lengthening telomeres) cell lines (McManus et al., 2009). Colorectal cancer cells with mutations in CDC4 or MRE11A were extremely susceptible to knockdown or chemical inhibition of FEN1 (van Pel et al., 2013). Although this study focused only on breast cancer, we believed that FEN1 inhibitor could be applied to treat a broad-spectrum of cancers.
Several attempts to develop FEN1 inhibitors have been described (Tumey et al., 2004, Tumey et al., 2005, Mcwhirter et al., 2013, Wadhwa and Jadhav, 2015, Dorjsuren et al., 2011). Tumey et al. reported that N-hydroxy urea derivatives are FEN1 inhibitors, and compound #20 has an impressive IC50 value of 3 nM (Tumey et al., 2005). To our knowledge, these are the most potent endonuclease inhibitors reported to date. These compounds sensitize cells to DNA damage-inducing agents, such as methyl methane sulfonate treatment. However, when used alone, its inhibitory effects on cell proliferation is low. Based on the structure of compound #20, we used pyrrolo[3.2-d]pyrimidine as a new pharmacophore and replaced the hydrophilic hydroxyl groups of urea with n-propyl building blocks. The resulting compound is more permeable to cells and introduces more cytotoxicity. For example, SC13 inhibits cell growth by > 20% at 5 μM, while the N-hydroxy urea derivatives inhibit cell growth by < 3% at the same concentration (Fig. 6a and data not shown).
We demonstrated that FEN1 is overexpressed in breast cancer cells and is essential for cancer cell proliferation. FEN1 enhances the resistance of cancer cells to chemotherapeutic agents that induce DNA lesions. Knockdown of FEN1 expression or SC13-mediated inhibition of FEN1 activity suppresses cell growth and sensitizes cancer cells to DNA damage-inducing drugs, both in cells and in an animal model. Further studies are underway to evaluate the clinical usefulness of this compound as a cancer therapy.
Funding Sources
This work was supported by the Ministry of Education, Science, and Technology of China [2013CB911600, NCET-13-0868], the National Natural Science Foundation of China [31271449], the Jiangsu Provincial Natural Science Foundation (BK20130044, BK20130061, BK2011783, BK20141448), CJ20160051. The Research Fund for the Doctoral Program of Higher Education of China (RFDP) (20133207110005), the Priority Academic Program Development of Jiangsu Higher Education Institutions (20110101), and the Project Sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (2015104SBJ0090).
Conflict of Interest Statement
We declare no financial or other relationships that may lead to a conflict of interest in this study.
Author Contributions
LF H. and Z G. and BH S. designed the study and supervised the entire project. YQ Z. designed the compound and HZ L. established the 3D model of SC13-FEN1 interaction. HY C. did the statistical analysis and database assay. F J. provided the cancer tissues and performed the IHC assay. HF S., YL Z., H Y. and H W. purified the proteins and performed all of the biochemical experiments in the current study. SC H. performed animal study. T Z., CS K. and XJ W. performed nuclease activity assay.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.11.012.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- Abdel-Fatah T.M., Russell R., Albarakati N., Maloney D.J., Dorjsuren D., Rueda O.M., Moseley P., Mohan V., Sun H., Abbotts R., Mukherjee A., Agarwal D., Illuzzi J.L., Jadhav A., Simeonov A., Ball G., Chan S., Caldas C., Ellis I.O., Wilson D.M., III, Madhusudan S. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol. Oncol. 2014;8:1326–1338. doi: 10.1016/j.molonc.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan L., Bambara R.A. Flap endonuclease 1. Annu. Rev. Biochem. 2013;82:119–138. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R., Kavuri S.M., Searleman A.C., Shen W., Shen D., Koboldt D.C., Monsey J., Goel N., Aronson A.B., Li S., Ma C.X., Ding L., Mardis E.R., Ellis M.J. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain M.C. Temozolomide: therapeutic limitations in the treatment of adult high-grade gliomas. Expert. Rev. Neurother. 2010;10:1537–1544. doi: 10.1586/ern.10.32. [DOI] [PubMed] [Google Scholar]
- Chung L., Onyango D., Guo Z., Jia P., Dai H., Liu S., Zhou M., Lin W., Pang I., Li H., Yuan Y.C., Huang Q., Zheng L., Lopes J., Nicolas A., Chai W., Raz D., Reckamp K.L., Shen B. The FEN1 E359K germline mutation disrupts the FEN1-WRN interaction and FEN1 GEN activity, causing aneuploidy-associated cancers. Oncogene. 2015;34:902–911. doi: 10.1038/onc.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorjsuren D., Kim D., Maloney D.J., Wilson D.M., III, Simeonov A. Complementary non-radioactive assays for investigation of human flap endonuclease 1 activity. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas A., Chovanec M. DNA double-strand break repair by homologous recombination. Mutat. Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Elakoum R., Gauchotte G., Oussalah A., Wissler M.P., Clement-Duchene C., Vignaud J.M., Gueant J.L., Namour F. CARM1 and PRMT1 are dysregulated in lung cancer without hierarchical features. Biochimie. 2014;97:210–218. doi: 10.1016/j.biochi.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Fehrmann R.S., Karjalainen J.M., Krajewska M., Westra H.J., Maloney D., Simeonov A., Pers T.H., Hirschhorn J.N., Jansen R.C., Schultes E.A., Van Haagen H.H., De Vries E.G., Te Meerman G.J., Wijmenga C., Van Vugt M.A., Franke L. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- Finger L.D., Blanchard M.S., Theimer C.A., Sengerova B., Singh P., Chavez V., Liu F., Grasby J.A., Shen B. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J. Biol. Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink D., Nebel S., Aebi S., Zheng H., Cenni B., Nehme A., Christen R.D., Howell S.B. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- Fink D., Aebi S., Howell S.B. The role of DNA mismatch repair in drug resistance. Clin. Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- Friedrich-Heineken E., Hubscher U. The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 2004;32:2520–2528. doi: 10.1093/nar/gkh576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Zhou H., Gan Y., Zhang J., Chen J., Gan X., Li H., Zheng W., Meng Z., Ma X., Wang X., Xu X., Xu G., Lu X., Liang Y., Zhang X., Huang W., Xu R. Small-molecule induction of phospho-eIF4E sumoylation and degradation via targeting its phosphorylated serine 209 residue. Oncotarget. 2015;6:15111–15121. doi: 10.18632/oncotarget.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn A., Stark J.M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- Guo Z., Qian L., Liu R., Dai H., Zhou M., Zheng L., Shen B. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol. Cell. Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zheng L., Dai H., Zhou M., Xu H., Shen B. Human DNA polymerase beta polymorphism, Arg137Gln, impairs its polymerase activity and interaction with PCNA and the cellular base excision repair capacity. Nucleic Acids Res. 2009;37:3431–3441. doi: 10.1093/nar/gkp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L. Breast cancer: challenges, controversies, breakthroughs. Nat. Rev. Clin. Oncol. 2010;7:669–670. doi: 10.1038/nrclinonc.2010.192. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C.A., Maitra A., Olsen M., Lowe A.W., Van Heek N.T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R., Jaffee E., Ryu B., Jones J., Eshleman J.R., Yeo C.J., Cameron J.L., Kern S.E., Hruban R.H., Brown P.O., Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A.S., Banerjee S., Aneja R., Sarkar F.H., Ostrov D.A., Narayan S. DNA polymerase beta as a novel target for chemotherapeutic intervention of colorectal cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B., Christmann M. DNA repair in resistance to alkylating anticancer drugs. Int. J. Clin. Pharmacol. Ther. 2002;40:354–367. doi: 10.5414/cpp40354. [DOI] [PubMed] [Google Scholar]
- Kao H.I., Henricksen L.A., Liu Y., Bambara R.A. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- Kim I.S., Lee M.Y., Lee I.H., Shin S.L., Lee S.Y. Gene expression of flap endonuclease-1 during cell proliferation and differentiation. Biochim. Biophys. Acta. 2000;1496:333–340. doi: 10.1016/s0167-4889(00)00029-x. [DOI] [PubMed] [Google Scholar]
- Krause A., Combaret V., Iacono I., Lacroix B., Compagnon C., Bergeron C., Valsesia-Wittmann S., Leissner P., Mougin B., Puisieux A. Genome-wide analysis of gene expression in neuroblastomas detected by mass screening. Cancer Lett. 2005;225:111–120. doi: 10.1016/j.canlet.2004.10.035. [DOI] [PubMed] [Google Scholar]
- Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M.F., Fan K., Russell R., Brown A.M., Kneitz B., Edelmann W., Kolodner R.D., Lipkin M., Kucherlapati R. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J.S., Seligson D.B., Yu H., Li A., Eeva M., Pantuck A.J., Zeng G., Horvath S., Belldegrun A.S. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006;98:445–451. doi: 10.1111/j.1464-410X.2006.06224.x. [DOI] [PubMed] [Google Scholar]
- Larsen E., Gran C., Saether B.E., Seeberg E., Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson M.H., Cummings N.M., Rassl D.M., Russell R., Brenton J.D., Rintoul R.C., Murphy G. Two novel determinants of etoposide resistance in small cell lung cancer. Cancer Res. 2011;71:4877–4887. doi: 10.1158/0008-5472.CAN-11-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen C.B., Neumayer L.A. Breast cancer: a review for the general surgeon. JAMA Surg. 2013;148:971–979. doi: 10.1001/jamasurg.2013.3393. [DOI] [PubMed] [Google Scholar]
- Mcmanus K.J., Barrett I.J., Nouhi Y., Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcwhirter C., Tonge M., Plant H., Hardern I., Nissink W., Durant S.T. Development of a high-throughput fluorescence polarization DNA cleavage assay for the identification of FEN1 inhibitors. J. Biomol. Screen. 2013;18:567–575. doi: 10.1177/1087057113476551. [DOI] [PubMed] [Google Scholar]
- Nguyen A., Ho L., Wan Y. Chemotherapy and Oncolytic Virotherapy: advanced tactics in the war against cancer. Front. Oncol. 2014;4:145. doi: 10.3389/fonc.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova T., Christmann M., Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 2009;29:2453–2459. [PubMed] [Google Scholar]
- Pascucci B., Russo M.T., Crescenzi M., Bignami M., Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 2005;33:280–288. doi: 10.1093/nar/gki168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E., Critchlow S.E., Teo S.H., Doherty A.J., Priestley A., Broughton B., Kysela B., Beamish H., Plowman N., Arlett C.F., Lehmann A.R., Jackson S.P., Jeggo P.A. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- Roos W.P., Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Sattler U., Frit P., Salles B., Calsou P. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep. 2003;4:363–367. doi: 10.1038/sj.embor.embor796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R.A., Holzen T.M. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Singh P., Liu R., Qiu J., Zheng L., Finger L.D., Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. BioEssays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- Singh P., Yang M., Dai H., Yu D., Huang Q., Tan W., Kernstine K.H., Lin D., Shen B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Nambiar M., Sharma S., Karki S.S., Goldsmith G., Hegde M., Kumar S., Pandey M., Singh R.K., Ray P., Natarajan R., Kelkar M., De A., Choudhary B., Raghavan S.C. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- Sun H., He L., Wu H., Pan F., Wu X., Zhao J., Hu Z., Sekhar C., Li H., Zheng L., Chen H., Shen B.H., Guo Z. The FEN1 L209P mutation interferes with long-patch base excision repair and induces cellular transformation. Oncogene. 2016 doi: 10.1038/onc.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresakumar P. San Jose State University; 2009. Use of Novel Assays to Measure in Vivo Base Excision DNA Repair. (Master's Theses. Paper 3681) [Google Scholar]
- Tomlinson G.E., Chen T.T., Stastny V.A., Virmani A.K., Spillman M.A., Tonk V., Blum J.L., Schneider N.R., Wistuba I.I., Shay J.W., Minna J.D., Gazdar A.F. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- Tsutakawa S.E., Classen S., Chapados B.R., Arvai A.S., Finger L.D., Guenther G., Tomlinson C.G., Thompson P., Sarker A.H., Shen B., Cooper P.K., Grasby J.A., Tainer J.A. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumey L.N., Huck B., Gleason E., Wang J., Silver D., Brunden K., Boozer S., Rundlett S., Sherf B., Murphy S., Bailey A., Dent T., Leventhal C., Harrington J., Bennani Y.L. The identification and optimization of 2.4-diketobutyric acids as flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 2004;14:4915–4918. doi: 10.1016/j.bmcl.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Tumey L.N., Bom D., Huck B., Gleason E., Wang J., Silver D., Brunden K., Boozer S., Rundlett S., Sherf B., Murphy S., Dent T., Leventhal C., Bailey A., Harrington J., Bennani Y.L. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 2005;15:277–281. doi: 10.1016/j.bmcl.2004.10.086. [DOI] [PubMed] [Google Scholar]
- Van Gent D.C., Hoeijmakers J.H., Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Van Pel D.M., Barrett I.J., Shimizu Y., Sajesh B.V., Guppy B.J., Pfeifer T., Mcmanus K.J., Hieter P. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa P.J.P., Jadhav H.R. QSAR and docking studies of N-hydroxy urea derivatives as flap endonuclease-1 inhibitors. Curr. Comput. Aided Drug Des. 2015;12:1. doi: 10.2174/1573409912666151124233628. [DOI] [PubMed] [Google Scholar]
- Wang K., Xie C., Chen D. Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis. Int. J. Mol. Med. 2014;33:1268–1274. doi: 10.3892/ijmm.2014.1682. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhou L., Li Z., Zhang T., Liu W., Liu Z., Yuan Y.C., Su F., Xu L., Wang Y., Zhou X., Xu H., Hua Y., Wang Y.J., Zheng L., Teng Y.E., Shen B. YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC Cancer. 2015;15:50. doi: 10.1186/s12885-015-1043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.G., Chang Y.L., Lin J.W., Wu C.T., Chen H.Y., Tsai M.F., Lee Y.C., Yu C.J., Shih J.Y. Including total EGFR staining in scoring improves EGFR mutations detection by mutation-specific antibodies and EGFR TKIs response prediction. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Qiu J., Huang Q., Shen B. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol. Cell. Biol. 2007;27:3176–3186. doi: 10.1128/MCB.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Zhou M., Li M., Singh P., Qiu J., Tsark W., Huang Q., Kernstine K., Zhang X., Lin D., Shen B. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- Zhou T., Pan F., Cao Y., Han Y., Zhao J., Sun H., Zhou X., Wu X., He L., Hu Z., Chen H., Shen B., Guo Z. R152C DNA Pol beta mutation impairs base excision repair and induces cellular transformation. Oncotarget. 2016;7:6902–6915. doi: 10.18632/oncotarget.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2