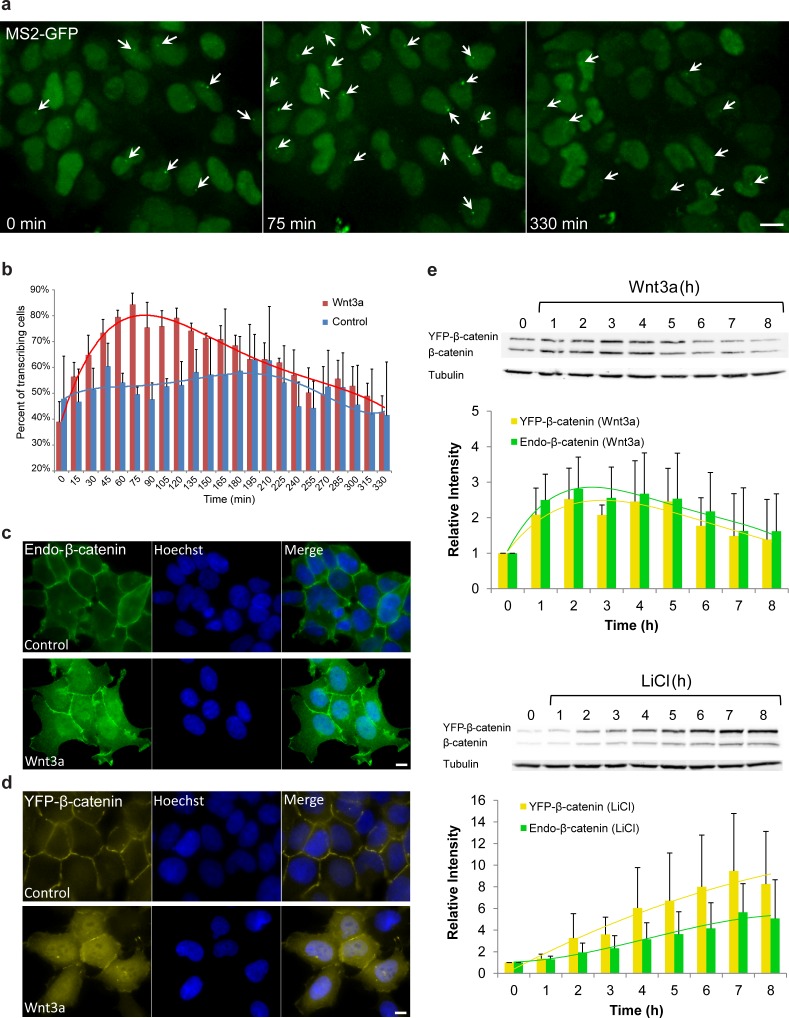
Figure 1. Cell system for following β-catenin intra-cellular dynamics and CCND1 transcription in single living cells.
(a) CCND1-MS2 HEK293 cells stably expressing MS2-GFP-CP were treated with Wnt3a and followed for 6 hr (every 15 min). Several frames from Video 1 are presented. The number of cells exhibiting transcriptionally active CCND1-MS2 genes (green dot in the nucleus, white arrow) was counted over time. Scale bar, 10 µm. (b) Plots showing the percentage of cells in the population with actively transcribing CCND1-MS2 genes in Wnt3a-treated (red, n = 98) and mock treated (blue, n = 128) cells. Mean ± sd from three fields imaged on different days—see Supplementary file 1a for statistics. (c) Immunofluorescence showing that endogenous β-catenin (green) is prominent at the cell membrane in untreated HEK293 cells (top) and accumulates in the cytoplasm and nucleus following activation by Wnt3a for 2 hr (bottom). Hoechst DNA stain is in blue. (d) Similar changes in the subcellular distribution following activation are seen in the YFP-β-catenin low-expressing clone. Bar = 10 μm. (e) Western blot time course of endogenous β-catenin and YFP-β-catenin protein accumulation following either Wnt3a (top) or LiCl (bottom) stimulation. Anti-β-catenin antibody was used for the detection of both β-catenin proteins. Tubulin was used as a loading control. Time 0 is the time point of activator addition. Blots are representative of 3 repeated experiments. The average quantification of 3 repeated experiments is presented in the plots below (mean ± sd). There is no statistical difference between the endogenous and exogenous levels of β-catenin in the two plots.