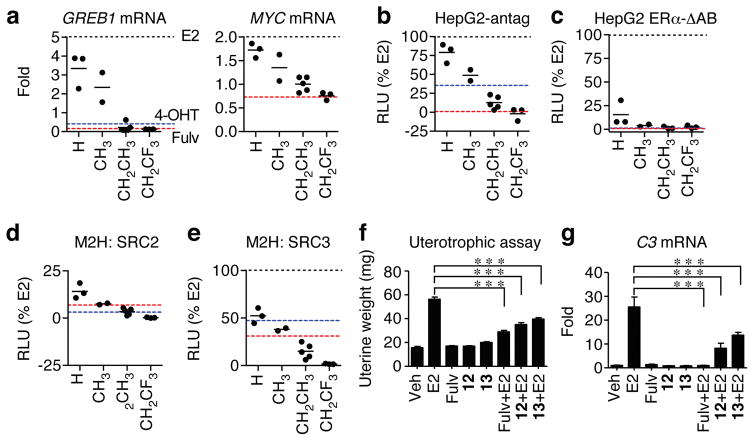
Figure 3. Modulation of ER α signaling by OBHS-N analogs in cells in vitro and in the mouse uterus in vivo.
(a–e) Dotted lines represent values for 10 nM E2 (black), 1 μM 4-OHT (cyan) and 1 μM fulvestrant (Fulv, red).
(a) Steroid-deprived MCF-7 cells were treated with 10 μM OBHS-N analogs for 24 hr. Total mRNA was reverse-transcribed and analyzed by qPCR for GREB1 and MYC expression relative to GAPDH. Values are mean, n = 2.
(b–c) Luciferase assays. HepG2 cells were co-transfected with 3xERE-luc reporter and ERα-WT or ERα-ΔAB expression plasmids were steroid-deprived and treated with 10 μM OBHS-N ligands. For the HepG2 antagonist mode assay, cells were cotreated with 10 nM E2. Values are mean, n = 3.
(d–e) HEK293T cells were co-transfected with a 5xUAS-luc reporter and expression plasmids for full-length ERα-VP16 and Gal4-SRC2/3 and were treated with 10 μM OBHS-N ligands. Values are mean, n = 3.
(f) Uterine weights of ovariectomized mice (n = 4 per group) treated for 4 days with vehicle (Veh), E2 (10 ng), fulvestrant (240 μg), or OBHS-N compounds 12 or 13 alone (240 μg), and in combination with E2. Values are mean ± s.e.m. Statistical significance was determined by ANOVA and Bonferroni’s Multiple Comparison test. *p < 0.05; **p < 0.01;***p < 0.001.
(g) Uteri (n = 4 per group) from animals in f were analyzed for expression the C3 gene. Values are mean ± s.e.m. Statistical significance was determined by ANOVA and Dunnett’s Multiple Comparison test. *p < 0.05; **p < 0.01;***p < 0.001.