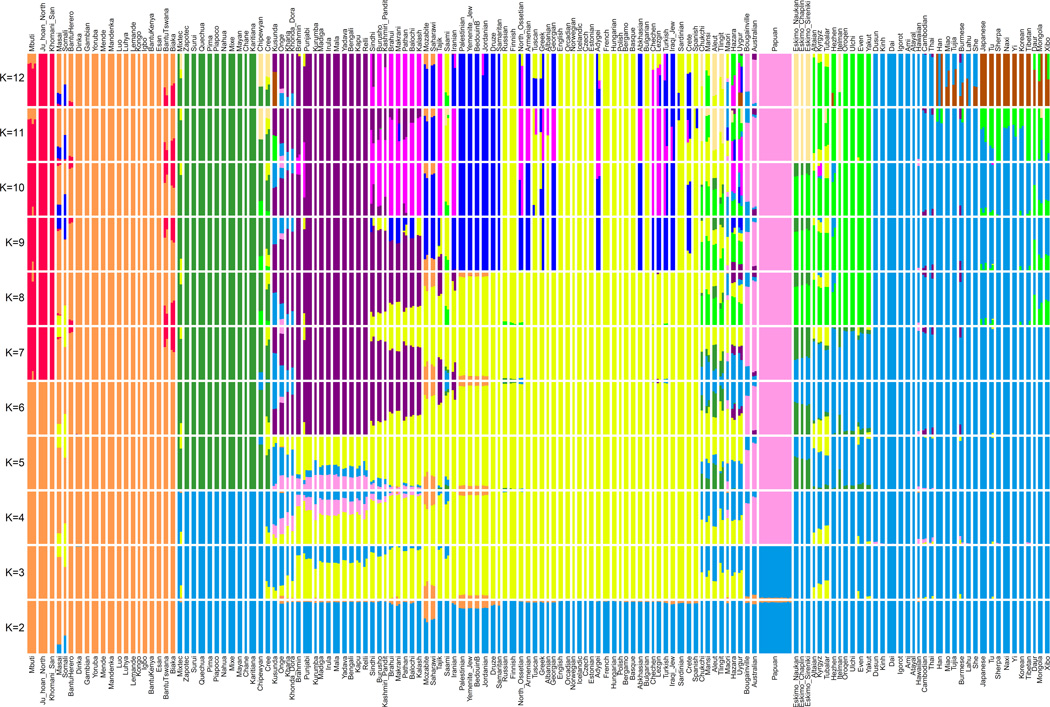
Extended Data Figure 3. ADMIXTURE analysis.
We carried out unsupervised ADMIXTURE 1.239,44 analysis over the 300 SGDP individuals in 20 replicates with randomly chosen initial seeds, varying the number of ancestral populations between K=2 and K=12 and using default 5-fold cross-validation (--cv flag). We used genotypes of at least filter level 1, and restricted analysis to sites where at least two individuals carried the variant allele (as singleton variants are non-informative for population clustering). After further filtering sites with at least 99% completeness and performing linkage-disequilibrium based pruning in PLINK 1.945,46 with parameters (--indep-pairwise 1000 100 0.2), a total of 482,515 single nucleotide polymorphisms remained. This figure shows the highest likelihood replicate for each value of K. We found that log likelihood monotonically increases with K, while the value K=5 minimizes cross-validation error (not shown). The solution at K=5 corresponds to major continental groups (Sub-Saharan Africans, Oceanians, East Asians, Native Americans, and West Eurasians), but we show the full range of K here as they illustrate finer-scale population structure that may be useful to users of the data.