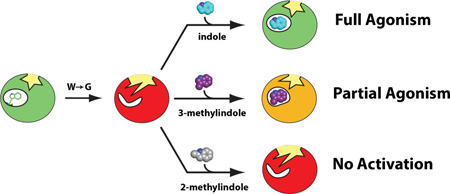
Abstract
Chemical biology has long sought to build protein switches for use in molecular diagnostics, imaging, and synthetic biology. The overarching challenge for any type of engineered protein switch is the ability to respond in a selective and predictable manner that caters to the specific environments and timescales needed for the application at hand. We previously described a general method to design switchable proteins, called “chemical rescue of structure”, that builds de novo allosteric control sites directly into a protein’s functional domain. This approach entails first carving out a buried cavity in a protein via mutation, such that the protein’s structure is disrupted and activity is lost. An exogenous ligand is subsequently added to substitute for the atoms that were removed by mutation, restoring the protein’s structure and thus its activity. Here, we begin to ask what principles dictate such switches’ response to different activating ligands. Using a redesigned β-glycosidase enzyme as our model system, we find that the designed effector site is quite malleable and can accommodate both larger and smaller ligands, but that optimal rescue comes only from a ligand that perfectly replaces the deleted atoms. Guided by these principles, we then altered the shape of this cavity by using different cavity-forming mutations, and predicted different ligands that would better complement these new cavities. These findings demonstrate how the protein switch’s response can be tuned via small changes to the ligand with respect to the binding cavity, and ultimately enabled us to design an improved switch. We anticipate that these insights will help enable design of future systems that tune other aspects of protein activity, whereby, like evolved protein receptors, remolding the effector site can also adjust additional outputs such as substrate selectivity and activation of downstream signaling pathways.
Graphical Abstract
Introduction
Chemical biology has been used to engineer small molecule-dependent activity into a variety of select proteins: this has allowed external activation of pathways controlling phosphorylation 1, glycosylation 2, and proteolysis 3, 4. However, each of these advances resulted from implementing pharmacological approaches uniquely tailored to the particular problem at hand. Recently, efforts have expanded to building synthetic switchable proteins in a more general way 5.
The potential utility of generalized protein switches is vast, and already they have successfully been used in certain cases to decipher cellular mechanisms and to construct novel devices such as biosensors 5–7. Some of these generalized examples include switches that activate protein function by using a small-molecule to reverse constitutive degradation of the target protein 3, 4 or induce removal of a self-splicing protein element (an “intein”) 8–10. These systems have typically been designed through an engineering-centric “bottom-up” approach 5: functional modules from different biological systems are mixed and matched to obtain the desired function. In another example, maltose-binding protein (input domain) was fused to β-lactamase (output domain) such that sugar binding regulated β-lactamase activity 11. However, engineering these switches is still far from rational; it entails extensive trial-and-error and/or directed evolution, particularly with regards to the linkers used for tethering together the functional modules 12. Individual point mutants can also tune the behavior of such protein switches, through effects on either the input domain (effector binding) and/or the output domain (catalysis) 13. While combinatorial libraries of randomized gene insertions and their point mutants can be tested to find combinations with the desired effect, it will nonetheless be advantageous to instead design these types of switches in a more rational way: this will enable the switch’s output(s) to not only be predicted, but also tuned for use in different environments and timescales.
As an alternative to this modular engineering strategy, we recently described a method called “chemical rescue of structure” that designs a new small molecule binding site directly into the protein’s output domain 14, 15. We start by identifying a “buttressing” side chain in the protein core (e.g. a tryptophan) that is required to maintain the architecture of the protein functional site; removal of this structural feature by mutation to glycine (e.g. W➝G) disrupts this architecture and leads to loss of function. We then restore the buttress by adding the cognate ligand (e.g. indole), which in turn restores the original protein conformation and rescues its activity.
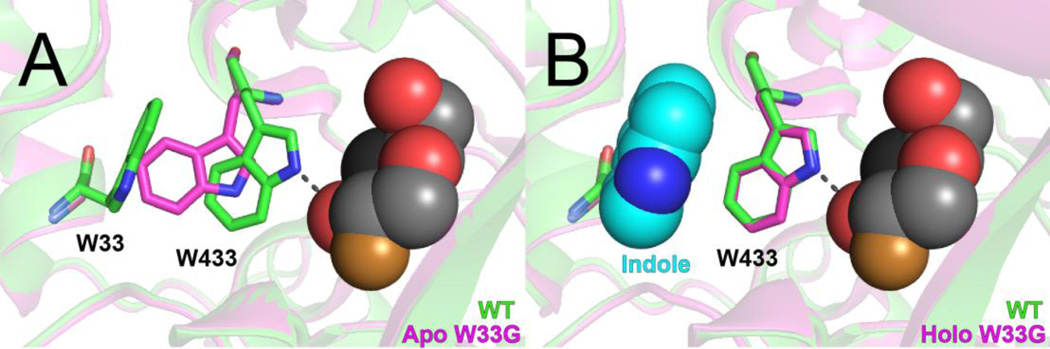
Our initial studies used β-glycosidase, a sugar-metabolizing enzyme from Sulfolobus solfataricus, as a model system. A high-resolution crystal structure revealed that when we introduced the W33G mutation, catalytic residue Trp433 shifted back into the newly-formed cavity (Figure 1a): this explained the observed loss of enzyme activity, because Trp433 engages in key hydrogen bonding interactions with the substrate. Strikingly, by solving a holo crystal structure we found that indole binding shifts Trp433 back to its former position in the wild type conformation (Figure 1b): this explained the observation that addition of indole restores enzyme activity. Using the Michaelis-Menten model to interpret the enzyme kinetics, we found that introduction of the W33G mutation led to a 730-fold decrease in kcat/Km relative to wild type; this was completely restored upon addition of 10 mM indole 14.
Figure 1. Structural basis for inactivation and rescue in β-gly W33G.
A comparison of the previously-determined crystal structures of wild type β-glycosidase (green) with its W33G mutant (pink). A covalent substrate analog (2-fluoro-2-deoxy-D-glucose, spheres) is included from WT structure to indicate the location of the active site; this compound was not present in either structure of β-gly W33G. (A) In the apo structure of β-gly W33G, W433 shifts into cavity that was previously occupied by W33; thus, W433 is no longer positioned to participate in hydrogen bonding with the substrate. (B) Upon soaking with indole, the holo structure of β-gly W33G shows that W433 has returned to its original position; indole (cyan spheres) occupies the cavity resulting from the W33G mutation.
Whether from “chemical rescue of structure” or from some other strategy, a current limitation in designing new molecular switches is the ability to selectively tune the responses of the switches in a predictable manner, as compared to the many other fields of engineering in which very precise control can be achieved 16. Biological systems are inherently noisy and complex, making it difficult to predict in a rational way how a given system will react to a particular input signal 17. In natural systems, such as G protein-coupled receptors (GPCRs), receptors have evolved to interact with a wide range of regulatory molecules 18 and can interact with different ligands that induce opposing responses 19. In estrogen receptor-α, ligand binding dynamics can dictate the fate of unique signaling pathways 20. In extreme cases, such as certain transcription factors, a single ligand can induce opposing responses on a given protein (agonist versus antagonist), depending on the context 21.
With regards to “chemical rescue of structure,” this comparison to natural signaling proteins prompts analogous questions. First, how should a cavity forming mutation be designed and then complemented in order to achieve optimal rescue? Second, how malleable is the resulting effector-binding site? In the case of the β-glycosidase example, the indole concentration can be varied in order to tune Vmax. But natural systems are rarely regulated by just one ligand – to what extent will this synthetic system respond to other ligands?
We envision two potential underlying structural mechanisms for rescue. In the first model the effector site behaves as a discrete switch, such that ligand binding exactly restores the geometry of the active site; such behavior is precedented by a recent study showing the discrete response of T4 lysozyme to a series of congeneric ligands 22. Given such a model, the degree to which a given ligand activates the protein would be related solely to the ligand’s binding affinity at the effector site. In the second model, the effector site responds in a continuous fashion: more diverse ligands can be accommodated, and each may influence protein structure – and thus activity – slightly differently. In this model, activation by a ligand is driven not just by its binding affinity, but also by the extent to which the ligand precisely restores the active site geometry for catalysis; analogous behavior is observed in crystal structures of GPCRs bound to full and partial agonists, in that the partial agonists make only a subset of the specific interactions of the full agonists 23.
In this study we seek to distinguish between these two models, and to further define the ideal chemical and geometric characteristics of a maximally rescuing ligand. We anticipate that the governing properties of this system will shed insight on the tunability of molecular switches built from “chemical rescue of structure” for future applications.
Results
Structure-activity relationship of alternate effector ligands
To probe the malleability of the effector site, we began by exploring the structure-activity relationship of rescue using a focused library of fifteen indole analogs with the β-glycosidase W33G enzyme switch described above. To probe the effector site cavity (the void left behind by the Trp to Gly mutation), we compiled this set by adding or removing functional groups around the indole parent scaffold. In order to explore in-plane substituents, methyl groups were individually added to each of the seven outer positions. To probe the effect of removing the hydrogen bond donor and allowing ring pucker, we tested indene and indan. We also included naphthalene and quinoline to explore the effect of increasing the ring size slightly, and we included benzene, toluene, and N-methylaniline to explore the effect of smaller compounds.
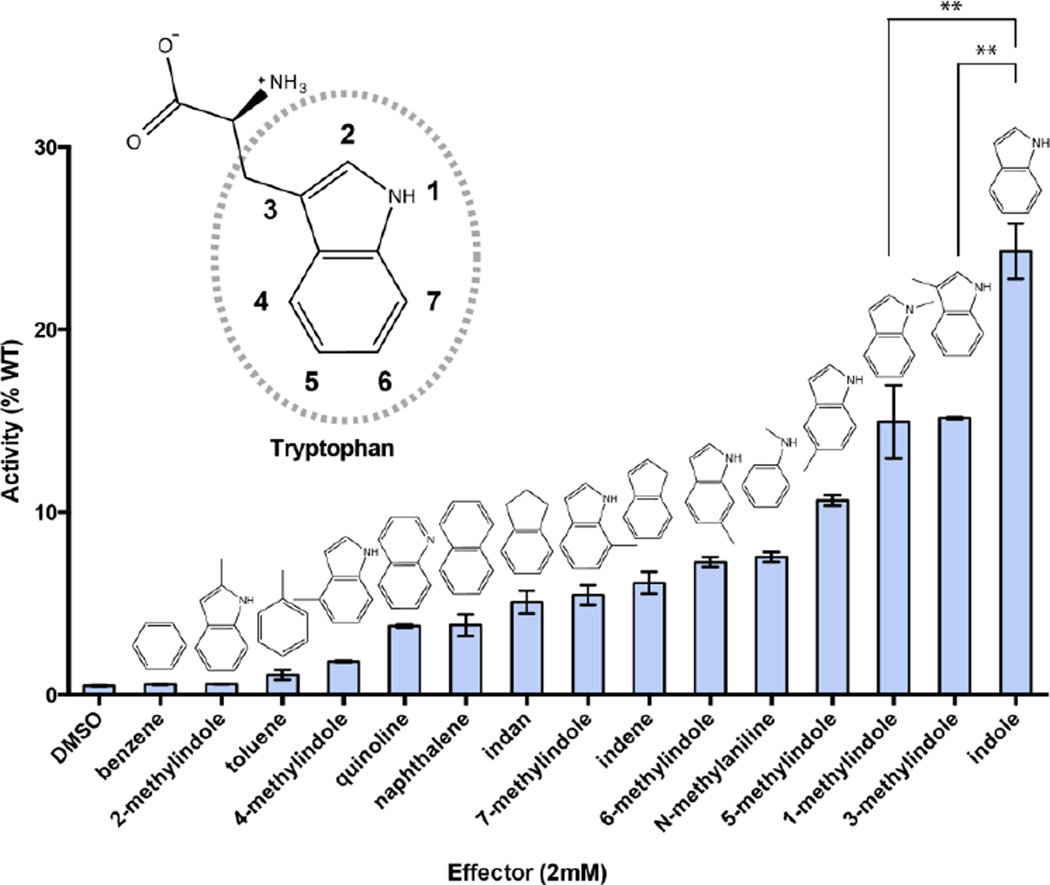
We tested each of these compounds in a β-glycosidase enzyme assay, to determine the extent to which each compound rescued activity (Figure 2). We used a high substrate concentration relative to the rescued enzymes’ Km values (Figure S1), such that our assay was designed to provide a readout on the effector’s influence on kcat. Because some effector compounds inhibited the wild type enzyme slightly at this high concentration, we normalized all results to the wild type enzyme under the same conditions. Each compound was tested at a concentration of 2 mM, because this was the maximum concentration at which all 15 compounds were soluble in our assay conditions.
Figure 2. Probing malleability of the β-gly W33G effector site.
The initial velocity is shown relative to the wild type enzyme in the presence of the same effector ligand; compounds are presented in ascending order of activity. Product formation was measured spectrofluorometrically using 750 μM fluorescein di-β-D-galactopyranoside (FDGal) as a substrate; each effector ligand was tested at 2 mM. All data were collected in triplicate, and error bars correspond to the standard error of the mean. Bars marked with ** have a statistically significant difference in their mean values (p < 0.01).
Relative to the wild type enzyme, W33G retains 0.5% basal activity in the absence of any ligand. In our previous study we found that indole fully restored wild type activity at a concentration of 10 mM 14; here, indole only restores 25% of the wild type activity because a lower concentration is used (2 mM). Of the 15 compounds tested, we found that 13 of them rescued activity; however, the extent of rescue was quite diverse. Among the seven methylindoles, for example, we observe the most rescue from 1- and 3-methylindoles; in contrast, 2-methylindole yields no increase in activity over the apo enzyme. Collectively these results show that structural details of the effector ligands can produce differing levels of catalytic activity – either through differences in binding affinity, or through conformational differences in the holo enzyme. In order to distinguish these possibilities, we next sought to directly detect binding of these ligands to the enzyme.
Direct determination of effector binding
We previously estimated the binding affinity of indole to β-gly W33G to be 0.75 mM in the presence of saturating substrate, and 15 mM in the absence of substrate 14. The weak binding affinity of this interaction – and presumably those of the indole analogs – places it outside the sensitivity limits of many common approaches for detecting protein-ligand interactions. To overcome this hurdle we therefore turned to 19F NMR 24, 25: the large chemical shift anisotropy of 19F nuclei results in line width differences of the fluorine signal between its free and protein-bound states, which is highly amenable for detection of weak interactions 26. In other words, differences between the transverse relaxation times of the free and protein-bound states are manifest through broadening of the 19F signal 26, 27.
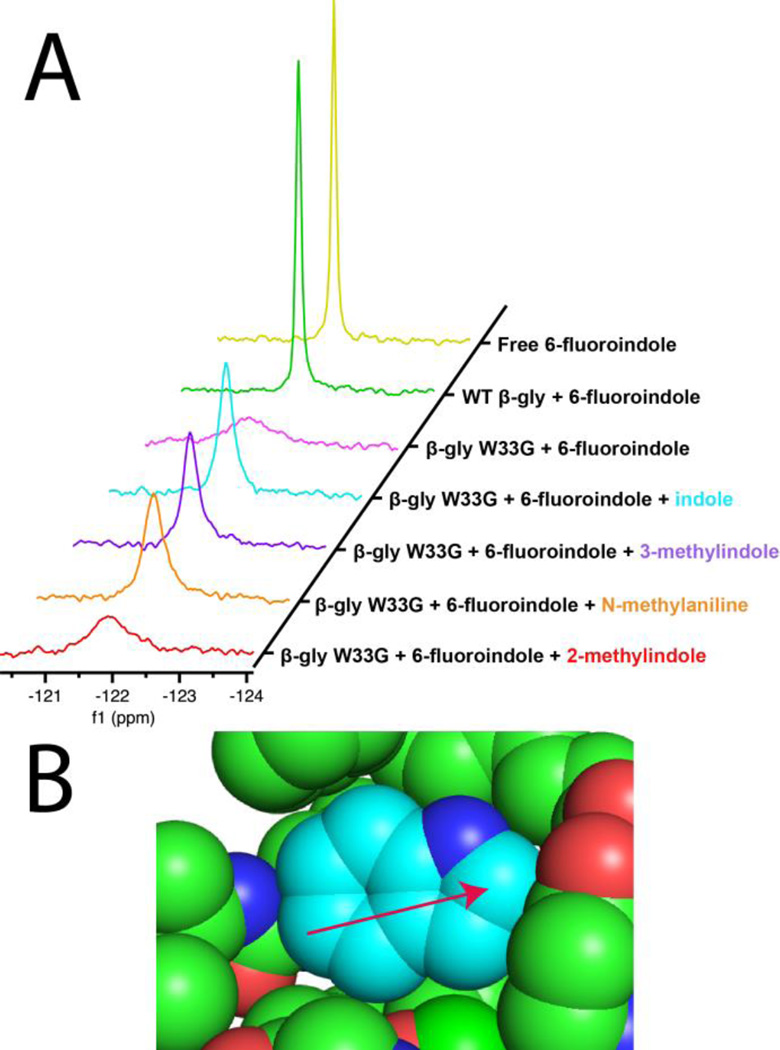
We began by identifying a fluorine-containing reporter molecule: we selected 6-fluoroindole, having found that this compound rescues enzyme activity at a very similar level as indole itself (Figure S2). Free in solution, the fluorine in this compound exhibits a sharp peak at −122.07 ppm. Upon addition of WT β-gly there is no change to this peak; however, upon addition of β-gly W33G we observe considerable peak broadening indicating that the reporter binds to the cavity-containing mutant (Figure 3a, Table S1). Importantly, for this experiment we included 2,4-dinitrophenyl 2-deoxy-2-fluoro-β-D-glucopyranoside, a covalently-attached substrate analog of this enzyme: given the allosteric linkage between the active site and effector site, we reasoned that inclusion of a substrate analog would order the active site residues for catalysis, and in doing so would also pre-order the effector site residues for ligand binding.
Figure 3. Direct determination of effector binding using a 19F NMR competition assay.
(A) 6-fluoroindole (2 mM) is used as a reporter molecule in all cases, with 500 μM protein concentration. Each competitor ligand was tested at a concentration of 5 mM. The sharp peak observed for the unbound reporter is broadened upon binding to the protein; ligands that compete for this binding site displace the reporter, leading to peak sharpening. (B) The packing of wild type β-glycosidase around Trp33 is tightest around the 2-position (arrow), making it particularly difficult to accommodate an extra methyl group here.
Next, we evaluated the ability of several ligands to compete with 6-fluoroindole for binding to β-gly W33G (presumably at the allosteric effector site). In addition to indole, we selected 3-methylindole (the next best rescuing compound), 2-methylindole (no rescue), and N-methylaniline (a smaller compound with intermediate rescue).
Upon addition of indole to our sample, we find that the 19F signal sharpens (Figure 3a, Table S1): this implies that indole displaces some of the 6-fluoroindole from the effector site. We also observe analogous peak sharpening for both 3-methylindole and N-methylaniline. In contrast, addition of 2-methylindole produces very little, if any, peak sharpening; this suggests that 2-methylindole binds to β-gly W33G much less strongly than 6-fluoroindole, and the other analogs included in this experiment.
This in turn provides an explanation for why 2-methylindole rescued activity less than other analogs (Figure 2). It is not the case that 2-methylindole binds to the allosteric effector site in a manner unproductive for catalysis; rather, 2-methylindole fails to rescue activity because it simply does not bind to this site. This explanation is further consistent with the protein structure (Figure 3b): the protein environment around Trp33 in the wild type enzyme is tightest around the 2-position, making this the most sterically challenging location at which to accommodate an extra methyl group.
19F NMR competition assays have been used to measure dissociation constants ranging from low nM to high μM 27, but unfortunately this experiment does not allow us to determine the binding affinities of these ligands in an accurate and quantitative way. Nonetheless, as a qualitative measure of binding this assay complements the enzymatic functional assay that will be presented below, for discriminating non-rescuing compounds that do not bind, versus those that bind but do not rescue.
Effect on rescue of both the substrate and the effector ligand
To directly examine potential differences in the active site geometry resulting from rescue by alternate ligands, we soaked crystals of β-gly W33G with 3-methylindole, with N-methylaniline, and with several other ligands. While soaking with indole led to a very useful crystal structure of that complex 14, upon soaking with other ligands we found only the unbound protein. Attempts to co-crystallize other ligands with β-gly W33G also yielded the unbound protein. For this reason, we sought to probe for small structural differences indirectly, through effects on substrate recognition.
Others have shown for allosteric activators of an unrelated system that the level of activation can depend on the structural properties of a specific substrate, and is thus a function of the precise arrangement of the activator-enzyme-substrate complex 28. Our previous study implicated Trp433 as the switch that distinguishes the active and inactive states of β-gly W33G. Accordingly, we anticipated that the precise placement of Trp433 in response to the rescuing ligand would be crucial for dictating the level of catalysis in the holo enzyme – and that rescue may be dependent not only on the effector ligand, but also on the choice of substrate.
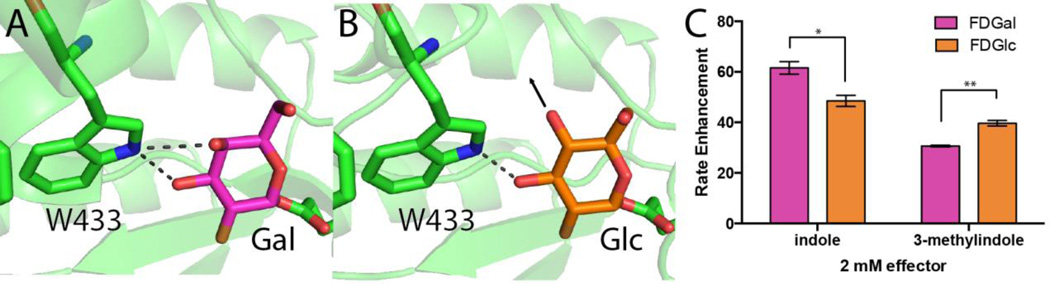
The structure of the wild type β-glycosidase has been solved in complex with two different covalent substrate analogs, 2-fluoro-2-deoxy-D-galactose (2F-Gal) and 2-fluoro-2-deoxy-D-glucose (2F-Glc) 29. While these two inhibitors form nearly identical interactions with the active site, the primary difference in these two structures is the interaction of the substrate analog with our “switch” residue, Trp433: 2F-Gal requires that the Trp433 sidechain splits its hydrogen bonding potential between the C3 and C4 hydroxyls (Figure 4a), whereas the different stereochemistry at the C4 position in 2F-Glc allows the Trp433 sidechain to hydrogen bond only with the C3 hydroxyl (Figure 4b). Based on this difference we anticipated that Gal-derived substrates would be more sensitive to structural perturbations of Trp433 than Glc-derived substrates. As a consequence, we further anticipated that Glc-derived substrates would be easier to rescue by effectors that do not perfectly fit the cavity, and may therefore not restore the perfectly ideal catalytic geometry of Trp433.
Figure 4. Trp433 forms different interactions with Gal versus Glc, allowing effectors to rescue to different extents, leading to slightly different effector preferences.
Crystal structures of wild type β-glycosidase bound to covalent substrate analogs (A) 2F-Gal and (B) 2F-Glc. The active site residue Trp433 makes distinct hydrogen bond interactions depending on the sugar’s stereochemistry at the C4 position. (C) Enhancement of initial velocity (relative to the apo enzyme) for substrates that differ only at this stereocenter (FDGal versus FDGlc), rescued using either indole or 3-methylindole. Assays were carried out using 250 μM substrate and 2 mM effector. Bars indicated have a statistically significant difference in their mean values (* with p < 0.05, ** with p < 0.01).
To test this hypothesis, we compared rescue using Gal-derived substrates versus Glc-derived substrates. Starting with the same Gal-derived substrate utilized in Figure 2 (fluorescein di-β-D-galactopyranoside, FDGal), we again observe better rescue with 2 mM indole than with 2 mM 3-methylindole. However, when we instead use the analogous Glc-derived substrate (fluorescein di-β-D-glucopyranoside, FDGlc) the difference in rescue between these substrates is greatly reduced (Figure 4c). As anticipated, then, rescue of the Gal-derived substrate depends more on precisely recapitulating the catalytic geometry of Trp433. If its position were perfectly superposed with the Trp33 sidechain in the wild type enzyme, the additional methyl group in 3-methylindole would align to the original Cβ position; in the rescued mutant, this would form a steric clash with the Cα to which it was previously covalently attached. We therefore expect that the 3-methylindole location would be slightly shifted relative to Trp33 (and relative to rescuing indole), which in turn explains its slightly decreased ability to rescue activity against FDGal. Though subtle, the ability of these rescuing ligands to shift the substrate preference for this enzyme is also highly reminiscent of “functional selectivity” in the pharmacology of natural signaling systems 30–33.
Separating effects of binding affinity from effects of active site geometry
To definitively separate effects of binding affinity from effects on the active site geometry, we next examined the concentration dependences of these effectors on the rate of product formation, using FDGal as substrate. To mitigate potential effects on Km (and thus simplify analysis), we again carried out experiments in a regime at which the substrate concentration (750 μM) is higher than Km of the rescued enzyme (Figure S1); thus, the observed initial velocity depends primarily on Vmax (which, in turn, depends in part on the effector concentration).
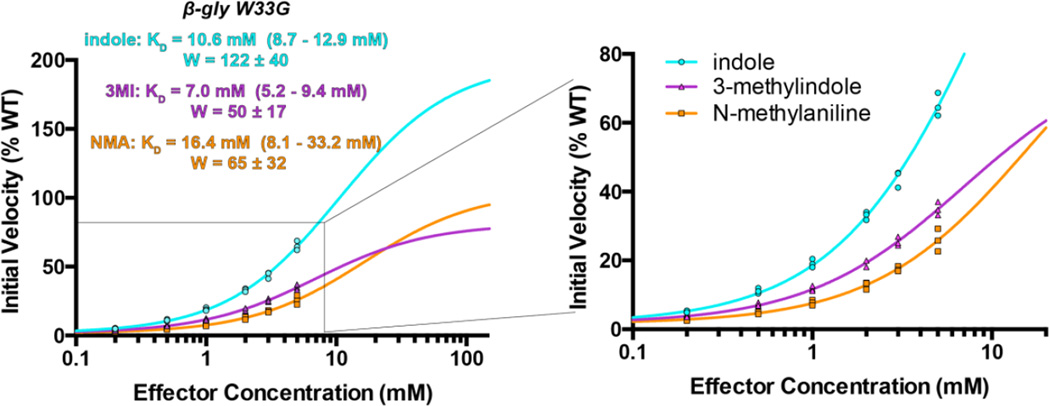
To fit the resulting initial velocities, we adapted the rate equation of a simple allosteric kinetic mechanism 34 by simplifying under the limit of saturating substrate (see Methods). This gave an equation that relates the change in initial velocities due to rescue to two key parameters, KD and W. KD is the effector dissociation constant, and W is a “linkage” term describing the magnitude of the effect of the allosteric ligand on Vmax. Structurally, the term W can also be interpreted as the extent to which the rescuing ligand restores the enzyme’s catalytic activity, because it reports on the rate of the holo enzyme.
Indole, 3-methylindole, and N-methylaniline each produce dose-dependent increases in the initial velocity that are well-described by this equation (Figure 5). To remain within the solubility limits for all three compounds, we were constrained to a maximum concentration of 5 mM. The curves have not fully saturated by this concentration, and so we present two views of the data: one that includes the extrapolated curve fitting (to show the full behavior of the model), and one that instead focuses only on the region for which experimental data was obtained (to show the agreement of the data to the model). Through these curve fits, the values of KD and W are obtained in tandem. To clearly establish the limits of this extrapolation, we report the bounds of KD and W (one standard error in either direction); the resulting bounds on the fitted curves are shown visually as well for comparison (Figure S3).
Figure 5. Full and partial agonism of β-gly W33G.
Enzyme assays carried out at high substrate concentration (750 μM FDGal) demonstrate the dependence of the initial velocity (and thus Vmax) on effector concentration. Both panels show the same data, expanded in the right panel to better show the agreement between the data and the model that was fit to it. All data were collected in triplicate, with all replicates shown explicitly on the plots. Experimental uncertainly of the fit values for KD and W are reported as a range corresponding to one standard error in either direction.
Among these three compounds, indole has the highest linkage term (W); this is consistent with our expectation that indole most effectively rescues the precise geometry of the active site. In fact, our extrapolation suggests that at high enough concentration the indole-rescued W33G mutant would actually surpass the rate of the wild type enzyme. While we cannot quite access these concentrations due to indole’s solubility, it is possible that the additional flexibility of the rescued enzyme allows W433G to adopt a conformation very slightly different than that of the wild type enzyme (and even more favorable for catalysis).
While the linkage term (W) for 3-methylindole is unsurprisingly worse than for indole, the KD value is actually slightly better than for indole. Thus, 3-methylindole appears to bind to β-gly W33G at least as tightly as indole, but does not rescue activity to the same extent because it does not fully restore the catalytic geometry. This observation is also consistent with those from the experiments described in the previous sections.
Finally, we compare the behavior of N-methylaniline to that of 3-methylindole. While the fitted values for W and KD are very similar for both ligands, the small differences provide an opportunity to visually highlight how these parameters together dictate the response to an effector ligand. 3-methylindole has a superior KD and thus more activity at low effector concentrations; in contrast, N-methylaniline has a superior W value and thus more activity at high effector concentrations – albeit at concentrations accessible only via extrapolation, due to solubility restrictions.
Drawing from long-established “receptor theory” pharmacology of natural signaling systems 35, 3-methylindole and N-methylaniline behave as partial agonists for the β-gly W33G “receptor”, in relation to the full agonist, indole.
Designing a better activating ligand for β-gly W33G
To test this model of β-gly W33G activation, we sought to rationally design a ligand with enhanced signaling relative to indole itself. Given our model, it will be very difficult to design a compound with more activity at high effector concentrations (better W), since this would require shifting the catalytic residues in a very prescribed way. Instead, we focused on ligands that might have better activity than indole at low effector concentrations, where binding affinity (better KD) is most responsible for determining activity.
Selectively adding fluorine is a common step in medicinal chemistry when seeking to improve potency of a compound: this makes the compound more lipophilic and also allows for productive halogen-pi interactions 36, 37. Because fluorine is isosteric with hydrogen, shape complementary for the receptor is typically preserved. Further, fluorine-substituted indoles (such as the reporter compound used in the 19F NMR competition assay) at almost all positions are commercially available.
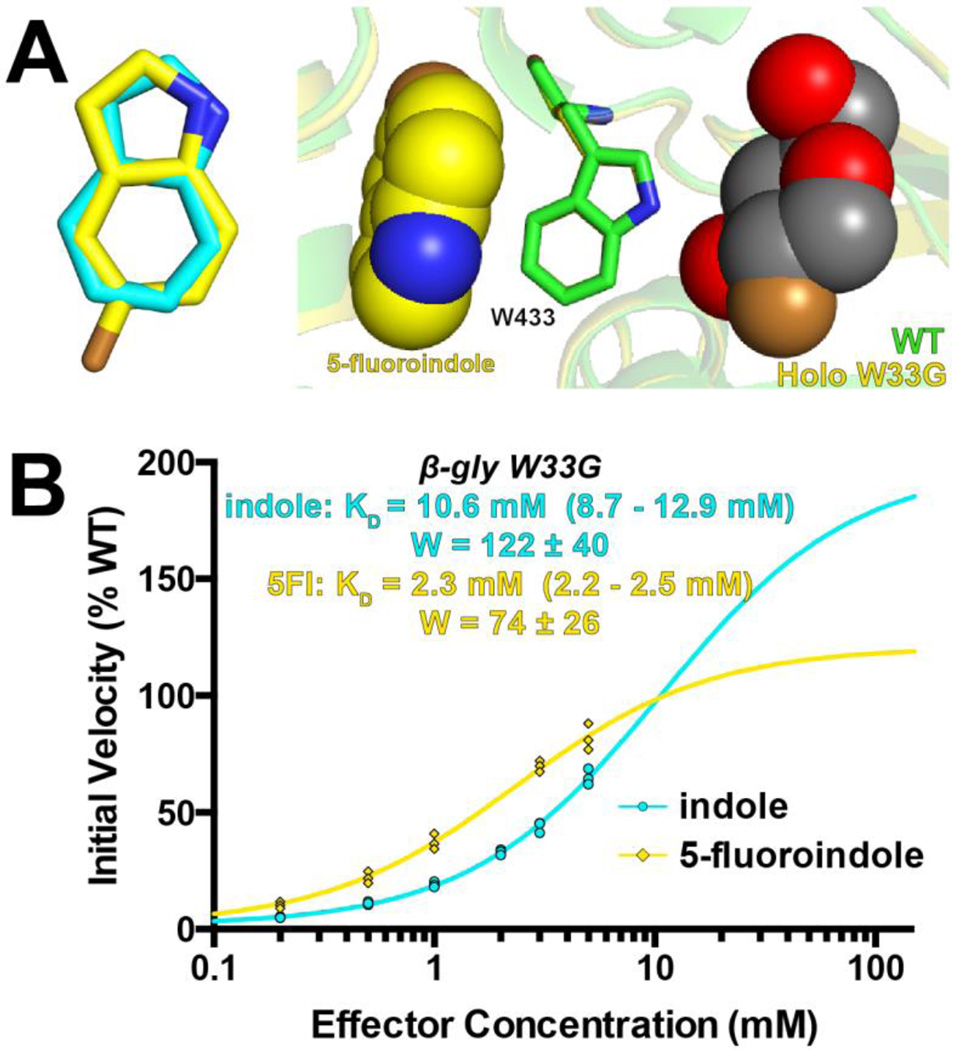
To begin, we solved the crystal structure of one of these, 5-fluoroindole, in complex with β-gly W33G (Figure 6a). Unsurprisingly, we found that 5-fluoroindole engages β-gly W33G in the same manner as indole, with no notable differences relative to the indole-bound or wild type structures of the enzyme. We then characterized rescue of activity from 5-fluoroindole, as we previously did for 3-methylindole and N-methylaniline. Indeed, relative to indole we find that 5-fluoroindole provides enhanced β-glycosidase activity at each of the concentrations we tested (Figure 6b). Notably, however, this difference is manifest most at low effector concentrations; as the concentration is increased indole starts to catch up with 5-fluoroindole, and is extrapolated to ultimately surpass the activity induced by 5-fluoroindole. This behavior is captured quantitatively through the parameters of each fit, in which KD favors 5-fluoroindole but W favors indole. While the crystal structure cannot provide an explanation for the difference in W, we attribute this to a very small difference in either the structure or dynamics of Trp433 in response to these two ligands.
Figure 6. 5-fluoroindole rescues β-gly W33G with a different signaling profile than indole.
(A) The crystal structure of 5-fluoroindole, solved to 1.75 Å resolution. At left, superposition with indole shows that 5-fluoroindole occupies essentially the same binding model (electron density for 5-fluoroindole is presented in Figure S4). At right, superposition with the wild type β-glycosidase shows that 5-fluoroindole restores the position of Trp433 (superposition with the indole-bound structure is presented in Figure S5). (B) β-gly W33G activity can be rescued either by indole or 5-fluoroindole. 5-fluoroindole binds more tightly, and thus rescues more effectively at low concentrations. At higher concentrations the difference in activity is reduced, and extrapolation suggests that indole will provide more activity than 5-fluoroindole at saturation. In all cases 750 μM FDGal was used as substrate. All data were collected in triplicate, with all replicates shown explicitly on the plots. Experimental uncertainly of the fit values for KD and W are reported as a range corresponding to one standard error in either direction.
Designing activating ligands for β-gly W33A
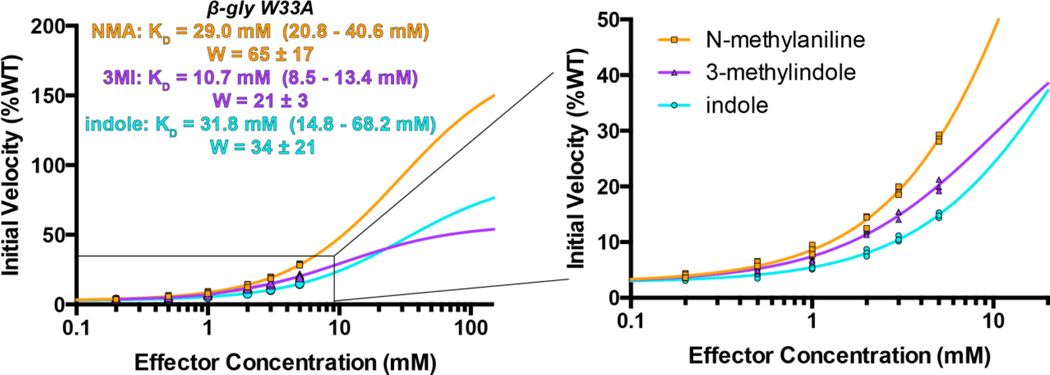
Encouraged by the ability to rationally design an altered response by varying the rescuing ligand, we next sought to adjust the shape of the β-gly cavity, and test whether complementary rescuing ligands could be designed to these new shapes. As a first experiment, we reduced the size of the cavity by incorporating W33A instead of W33G. Earlier, we explained the preference of β-gly W33G for indole over 3-methylindole (Figure 5) by pointing out that while 3-methylindole restores the same number of removed (non-hydrogen) atoms that were removed by mutation, rescue would require close contact between non-covalent atoms that were previously bonded to one another (Cα and Cβ). Applying this logic to W33A, then, one would expect a steric clash between the Cβ of Ala33 and the atom replacing the Cγ of the original tryptophan sidechain (i.e. the indole 3-position). Instead, one would predict better rescue by the corresponding compound that lacks this atom: N-methylaniline. Spurred by this rationale, we therefore explored rescue of β-gly W33A by N-methylaniline, indole, and 3-methylindole.
Consistent with this expectation, we find that N-methylaniline rescues β-gly W33A better than either indole or 3-methylindole (Figure 7); however, the difference was surprisingly modest. A further surprise was the fact that 3-methylindole rescued activity better than indole, which was counterintuitive given that this experiment entailed reducing the size of the cavity.
Figure 7. N-methylaniline is the optimal rescuing ligand for β-gly W33A.
The relatively modest rescue by N-methylaniline, as well as the fact that 3-methylindole rescued better than indole, can be explained by separately examining the binding affinity (KD) and contribution to the active site geometry (W) for each compound. In all cases 750 μM FDGal was used as substrate. All data were collected in triplicate, with all replicates shown explicitly on the plots. Experimental uncertainly of the fit values for KD and W are reported as a range corresponding to one standard error in either direction.
The explanation for both of these puzzles is found in the separate contributions from active site geometry (W) and binding affinity (KD). N-methylaniline was expected to be most compatible with the Trp433 geometry that is optimal for catalysis; indeed, N-methylaniline has the highest value of W. Its relatively modest activation, meanwhile, derives solely from its poor binding affinity, which may reflect the entropic cost of ordering its freely rotatable bond (that is not present in the other compounds). In the same vein, the surprisingly good activity of 3-methylindole relative to indole also derives exclusively from a difference in binding affinity, presumably because 3-methylindole is more hydrophobic than indole. While it remains surprising that the value of W is not much worse for 3-methylindole than for indole, we speculate that 3-methylindole may flip to position its methyl group towards the exposed region used by indole’s amine group (Figure 3b); this hypothesis could best be tested through crystallographic evidence, however we have been unable to solve a structure of either β-gly mutant in complex with 3-methylindole. Finally, we note that the 3-methylindole and indole values of W are lower for W33A than for W33G; this is also consistent with the hypothesis that neither of the compounds can restore the active site structure quite as accurately when disrupted by Ala33.
In summary, while N-methylaniline proved to be the optimal ligand for rescuing this smaller cavity, the magnitude to which catalytic activity could be restored was nonetheless limited by the small size and flexible nature of this compound. To design a more sensitive switch, we therefore introduced a larger cavity into this enzyme.
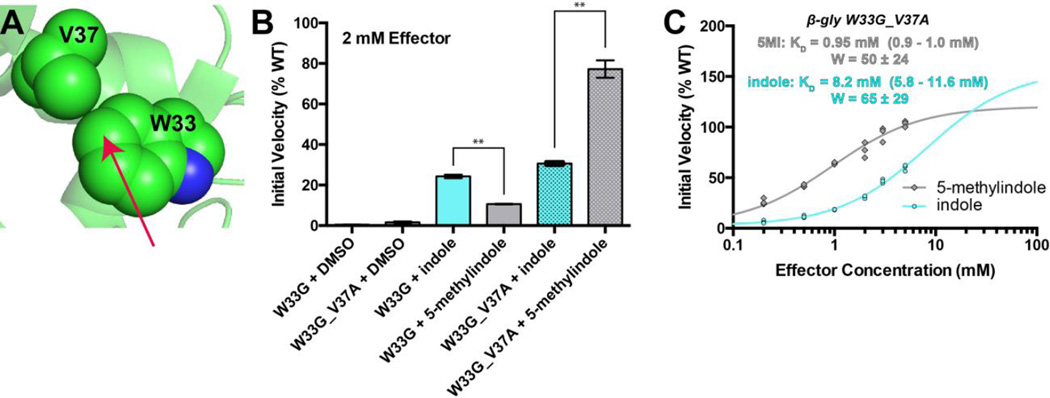
Designing activating ligands for β-gly W33G_V37A
Based on the structure of the wild type enzyme, there was only a single nearby sidechain in plane with the indole ring of Trp33, Val37 (Figure 8a). To expand the allosteric effector site, we therefore introduced an additional mutation, V37A, into β-gly W33G. Based on the location of this additional cavity-forming mutation, we expected that β-gly W33G_V37A would best be rescued by indole substituted at the 5-position.
Figure 8. 5-methylindole effectively rescues β-gly W33G_V37A.
(A) Based on the crystal structure of the wild type enzyme, Val37 was a natural location at which to expand the allosteric effector site. We anticipated that reducing the size of this sidechain would allow an additional substituent at indole’s 5-position (arrow). (B) At a concentration of 2 mM, indole rescues the β-gly W33G and W33G_V37A mutants about equally well. Relative to indole, 5methylindole rescues β-gly W33G much less, and β-gly W33G_V37A much more. Error bars correspond to the standard error of the mean, and bars marked with ** have a statistically significant difference in their mean values (p < 0.01). (C) By separately examining the binding affinity (KD) and contribution to the active site geometry (W), it is clear that the basis for the difference in rescue of W33G_V37A derives from the tighter binding affinity of 5-methylindole for this double mutant. In all cases 750 μM FDGal was used as substrate. All data were collected in triplicate, with all replicates shown explicitly on the plots. Experimental uncertainly of the fit values for KD and W are reported as a range corresponding to one standard error in either direction.
We initially tested the activity of both indole and 5-methylindole against β-gly W33G, at a concentration of 2 mM (Figure 8b). As in our previous experiment (Figure 2), we found that indole conferred about twice the activity of 5-methylindole under these conditions. Testing these two compounds against β-gly W33G_V37A, however, showed a reversal of their activities: now 5-methylindole proved superior over indole.
To understand the basis for this difference, we explored β-gly W33G_V37A activity as a function of effector concentration (Figure 8c). Perhaps unsurprisingly, both compounds have very similar values of W: this implies that both compounds engage the effector site with very similar binding modes, and thus interact with Trp433 in the same way. Accordingly, the basis for the greater potency of 5-methylindole comes from its binding affinity (KD), which is almost an order of magnitude tighter than that of indole. This observation is also unsurprising, given the extra hydrophobic surface area that is buried upon binding of 5-methylindole instead of indole.
At the outset of this study, we tested a panel of indole analogs at 2 mM for rescue of β-gly W33G, and found that none of these surpassed the 25% of wild type activity recovered by indole itself (Figure 2). Now, with a more refined view of this system predicated on pharmacological receptor theory 35, we have designed a new protein-ligand pair that exhibits almost 80% of wild type activity at 2 mM effector (Figure 8b).
Discussion
In the introduction to this study, we laid out two potential structural mechanisms for rescue at this designed allosteric effector site. In the first model the effector site behaves as a discrete active/inactive switch, activated through ligand binding. In the second model the effector site behaves as a rheostat, reporting not only on the presence of an activating ligand, but also on its precise structural complementarity for the effector site.
All of the accumulated evidence in our study points to the latter model. Numerous methylindoles show activity (all but 2-methylindole), highlighting the malleability of this site for accommodating alternate ligands. While certain ligands bind to this site more tightly than indole, none rescue the precise active site geometry optimal for catalysis quite as effectively as indole: instead, they serve as partial agonists of β-gly W33G.
Through the structure-activity relationship compiled for β-gly W33G, two principles emerge that guide design of agonists to fit a given cavity. First, the ideal rescuing ligand should replace the protein atoms removed by mutation as closely as possible: this leads to the most activity once the ligand is bound. Second, there may be a small tradeoff between a steric clash and a small cavity when replacing the deleted atoms: as demonstrated through indole’s rescue of β-gly W33G and N-methylaniline’s rescue of β-gly W33A, the small unfilled volume is less detrimental for activity than the structural rearrangement required to resolve the steric clash. With these design principles in mind, it became possible to rationalize the complete behavior of many different activating ligands, and also to design a new switch with enhanced activity.
The key strategy motivating design of the 5-methylindole / β-gly W33G_V37A switch was to improve binding affinity while at the same time maintaining the precise complementarity that restored the optimal geometry of Trp433. Given the very small ligands (indole has only 9 non-hydrogen atoms), there is an intrinsic biophysical limit to the potential binding affinity that can be achieved 38, 39. Indeed, the ligand efficiency (binding free energy per non-hydrogen atom) 38 of indole binding to β-gly W33G is −0.31 kcal/mol·atom (Table S2), which is comparable to the median ligand efficiency of other protein-ligand complexes (−0.34 kcal/mol·atom) 39. By design, 5-methylindole incorporates an additional potent interaction (burial of the methyl group in a tight cavity), increasing the ligand efficiency to −0.43 kcal/mol·atom. Given the typical limits of ligand efficiency 39, designing switches that respond to lower ligand concentrations will likely require the use of ligands that rescue larger cavities, or more sophisticated mechanisms that incorporate feed-forward signaling.
In our studies of other model systems activated by indole after incorporation of W➝G cavity-forming mutations, we found that inactivation and rescue could be mediated by local or global unfolding/refolding, rather than through a discrete conformational change 15. This mechanism enabled the use of effector sites located much further from the active site, but may come at the expense of sensitivity towards slightly different ligands. Here, we demonstrated that the response to different ligands could be tuned through a combination of binding affinity (KD) and precise structural complementarity (W). We anticipate that longer-ranged allosteric switches built in manner are less likely to preserve the subtle structural response to different ligands as the signal is transduced through the protein; as a result, the response may instead be dominated simply by the binding affinity. The same limitation is also likely to switches that are built by recombining modular domains 40, 41, because it is difficult to predict precisely how the linkers will transduce the input signal over to the output domain. An important advantage of chemical rescue of structure is the ability to place the location of the effector site in close proximity to the output signal (i.e. the active site), which in turn may facilitate the graded responses we demonstrate here.
Looking forward, careful modulation of the response over a short distance may also facilitate design of switches that exhibit functional selectivity. Already we observe this property in β-gly W33G – though not explicitly designed – in the fact that indole and 3-methylindole tune the enzyme’s preference for substrates containing galactose versus glucose. Natural systems make extensive use of this paradigm, as exemplified by GPCRs that initiate signaling through either the canonical G protein-mediated pathway or the non-canonical β-arrestin G protein-independent pathway, depending on the particular ligand that it bound 19, 42, 43. We anticipate that the design principles emerging from this study of the β-gly W33G designed allosteric effector site will provide a first step towards rationally designing new synthetic switches capable of absorbing information from several different molecular cues, and providing distinct and meaningful responses to each of them 5.
Methods
A complete description of methods is available as Supporting Online Materials. Coordinates and structure factors for the crystal structure of β-gly W33G bound to 5-fluoroindole has been deposited with the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) with accession code 5IXE.
19F NMR competition assay
The weak affinity of the interactions we sought to measure places them outside the sensitivity limits of techniques to such as surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), and differential scanning fluorimetry (DSF). The sensitivity problem is exacerbated for some of these techniques by the particularly small ligands (< 200 Da) and the relatively large protein (56,000 Da). We recognized that the problem of detecting very weak binding of a small ligand is reminiscent of the challenges faced in fragment-based drug discovery campaigns, and therefore borrowed an emerging tool from their repertoire, 19F NMR 24, 25.
19F NMR spectra were acquired on a Bruker DRX spectrometer equipped with an 11.7 T magnet (19F resonance frequency equals 470 MHz). 500 μM β-glycosidase in 50 mM phosphate buffer in H2O with 10% protonated DMSO, 2 mM 6-fluoroindole and 5 mM of the competitor ligand.
Protein samples were pre-treated with 2,4-dinitrophenyl 2-deoxy-2-fluoro-β-D-glucopyranoside. The 2,4-dinitrophenol serves as a leaving group such that the 2-deoxy-2-fluoro-β-D-glucose remains covalently attached to the protein. We confirmed labeling of the protein by broadening of the inhibitor’s 19F NMR peak, and also spectrophotometrically by production of 2,4-dinitrophenol.
β-glycosidase enzyme assay
All β-glycosidase enzyme assays were conducted using 58 nM β-glycosidase with fluorescein di-β-D-galactopyranoside (FDGal) or fluorescein di-β-D-glucopyranoside (FDGlc) as substrate, in a buffer of 50 mM sodium phosphate pH 6.5 and 10% DMSO at 37°C. Upon catalysis, FDGal is cleaved twice yielding one D-galactose and two molecules of fluorescein. We detect product formation by following fluorescence with excitation at 485 nm and emission at 528 nm.
Rate equation
To fit the observed initial velocities, we adapted the rate equation of a simple allosteric kinetic mechanism 34 by simplifying under the limit of saturating substrate. This gave the relationship:
| (Equation 1) |
where V is the initial velocity at a given effector concentration, [A] is the effector concentration, KD is the effector dissociation constant, Vapo is the maximal velocity in the absence of effector, and W is a “linkage” term describing the magnitude of the effect of the allosteric ligand on Vmax. Functionally, W is defined as the ratio of the maximal velocity at saturating effector, to the maximal velocity in the absence of effector (in other words, W is the fold-increase in Vmax that the effector can bring about). Throughout this work we interpret W as the extent to which the rescuing ligand restores the protein structure required for catalysis, because it reports on the rate of the holo enzyme.
Statistical Analysis
Statistical significance of differences in the standard error of the mean were evaluated using two-tailed t-tests (Figures 2, 4, and 8).
Standard errors for log(KD) and W were calculated with GraphPad Prism version 6.0 via:
| (Equation 2) |
where Pi represents the ith adjustable (non-constant) parameter. SS is the sum of the squared residuals. DF is the degrees of freedom (the difference between the number of data points and parameters fit by regression). Cov(i,i) is the i-th diagonal element of the covariance matrix.
The standard error of log(KD) was used to calculate the upper and lower bounds of the KD values (one standard error above/below the best-fit value).
Supplementary Material
Acknowledgments
We thank Andrea Bazzoli, Yan Xia, Jittasak Khowsathit, Andrew Beaven, and Wonpil Im for valuable discussions and feedback. Support for the NMR instrumentation was provided by NSF Major Research Instrumentation Grant 9977422 and NIH Center Grant 5P20GM103418. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was supported by grants from the National Institute of General Medical Sciences (8P20GM103420 and 5P30GM110761), the National Center for Research Resources (5P20RR017708), the National Science Foundation through XSEDE allocation MCB130049, the Human Frontier Science Program (J.K.), and the Alfred P. Sloan Fellowship (J.K.). S. Jimmy Budiardjo was supported by a Post Baccalaureate Research Education grant (2R25GM078441) and the Pharmaceutical Aspects of Biotechnology training grant (5T32GM008359) from the National Institutes of General Medical Sciences of the National Institutes of Health. Timothy J. Licknack was supported by the NSF REU Program (DBI-1156856). Michael B. Cory was supported by the Beckman Scholars Award from the the Arnold and Mabel Beckman Foundation.
Footnotes
Associated Content (Supporting Information)
A complete description of experimental methods and procedures. Figure S1 showing Km values for W33G when rescued by three different effectors. Figure S2 showing indole and 6-fluoroindole activity for β-gly W33G. Figure S3 showing upper and lower bounds of initial velocity at ± 1 standard error. Figure S4 showing electron density maps of 5-fluoroindole bound to β-gly W33G. Figure S5 showing superposition of 5-fluoroindole-bound and indole-bound crystal structures. Table S1 containing 19F NMR peak integrals. Table S2 containing ligand efficiency values for each ligand/protein pair reported in this study. Table S3 containing crystallographic data for 5-fluoroindole-bound β-gly W33G. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28:743-U1756. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czlapinski JL, Schelle MW, Miller LW, Laughlin ST, Kohler JJ, Cornish VW, Bertozzi CR. Conditional glycosylation in eukaryotic cells using a biocompatible chemical inducer of dimerization. J Am Chem Soc. 2008;130:13186–13187. doi: 10.1021/ja8037728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pratt MR, Schwartz EC, Muir TW. Small-molecule-mediated rescue of protein function by an inducible proteolytic shunt. Proc Natl Acad Sci U S A. 2007;104:11209–11214. doi: 10.1073/pnas.0700816104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein V, Alexandrov K. Synthetic protein switches: design principles and applications. Trends Biotechnol. 2015;33:101–110. doi: 10.1016/j.tibtech.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Golynskiy MV, Koay MS, Vinkenborg JL, Merkx M. Engineering protein switches: sensors, regulators, and spare parts for biology and biotechnology. Chembiochem. 2011;12:353–361. doi: 10.1002/cbic.201000642. [DOI] [PubMed] [Google Scholar]
- 7.Merkx M, Golynskiy MV, Lindenburg LH, Vinkenborg JL. Rational design of FRET sensor proteins based on mutually exclusive domain interactions. Biochem Soc Trans. 2013;41:1201–1205. doi: 10.1042/BST20130128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buskirk AR, Ong YC, Gartner ZJ, Liu DR. Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc Natl Acad Sci U S A. 2004;101:10505–10510. doi: 10.1073/pnas.0402762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci U S A. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiraga K, Soga I, Dansereau JT, Pereira B, Derbyshire V, Du Z, Wang C, Van Roey P, Belfort G, Belfort M. Selection and structure of hyperactive inteins: peripheral changes relayed to the catalytic center. J Mol Biol. 2009;393:1106–1117. doi: 10.1016/j.jmb.2009.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke W, Laurent AH, Armstrong MD, Chen Y, Smith WE, Liang J, Wright CM, Ostermeier M, van den Akker F. Structure of an engineered beta-lactamase maltose binding protein fusion protein: insights into heterotropic allosteric regulation. PLoS One. 2012;7:e39168. doi: 10.1371/journal.pone.0039168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci U S A. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JH, Xiong T, Ostermeier M. The interplay between effector binding and allostery in an engineered protein switch. Protein Sci. 2016 doi: 10.1002/pro.2962. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckert K, Budiardjo SJ, Brunner LC, Lovell S, Karanicolas J. Designing allosteric control into enzymes by chemical rescue of structure. J Am Chem Soc. 2012;134:10055–10060. doi: 10.1021/ja301409g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Y, DiPrimio N, Keppel TR, Vo B, Fraser K, Battaile KP, Egan C, Bystroff C, Lovell S, Weis DD, Anderson JC, Karanicolas J. The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation. J Am Chem Soc. 2013;135:18840–18849. doi: 10.1021/ja407644b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ang J, Harris E, Hussey BJ, Kil R, McMillen DR. Tuning Response Curves for Synthetic Biology. ACS Synth Biol. 2013;2:547–567. doi: 10.1021/sb4000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy KF, Adams RM, Wang X, Balazsi G, Collins JJ. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 2010;38:2712–2726. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nat Chem Biol. 2012;8:670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbas A, Roth BL. Arresting serotonin. Proc Natl Acad Sci U S A. 2008;105:831–832. doi: 10.1073/pnas.0711335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan S, Nwachukwu JC, Parent AA, Cavett V, Nowak J, Hughes TS, Kojetin DJ, Katzenellenbogen JA, Nettles KW. Ligand-binding dynamics rewire cellular signaling via estrogen receptor-alpha. Nat Chem Biol. 2013;9:326–332. doi: 10.1038/nchembio.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motlagh HN, Hilser VJ. Agonism/antagonism switching in allosteric ensembles. Proc Natl Acad Sci U S A. 2012;109:4134–4139. doi: 10.1073/pnas.1120519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merski M, Fischer M, Balius TE, Eidam O, Shoichet BK. Homologous ligands accommodated by discrete conformations of a buried cavity. Proc Natl Acad Sci U S A. 2015;112:5039–5044. doi: 10.1073/pnas.1500806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AG, Schertler GF, Tate CG. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalvit C. NMR methods in fragment screening: theory and a comparison with other biophysical techniques. Drug Discov Today. 2009;14:1051–1057. doi: 10.1016/j.drudis.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Jordan JB, Poppe L, Xia X, Cheng AC, Sun Y, Michelsen K, Eastwood H, Schnier PD, Nixey T, Zhong W. Fragment based drug discovery: practical implementation based on (1)(9)F NMR spectroscopy. J Med Chem. 2012;55:678–687. doi: 10.1021/jm201441k. [DOI] [PubMed] [Google Scholar]
- 26.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Curr Top Med Chem. 2003;3:39–53. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- 27.Dalvit C, Fagerness PE, Hadden DT, Sarver RW, Stockman BJ. Fluorine-NMR experiments for high-throughput screening: theoretical aspects, practical considerations, and range of applicability. J Am Chem Soc. 2003;125:7696–7703. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- 28.Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, Stein RL. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gloster TM, Roberts S, Ducros VM, Perugino G, Rossi M, Hoos R, Moracci M, Vasella A, Davies GJ. Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors. Biochemistry. 2004;43:6101–6109. doi: 10.1021/bi049666m. [DOI] [PubMed] [Google Scholar]
- 30.Johnson EA, Oldfield S, Braksator E, Gonzalez-Cuello A, Couch D, Hall KJ, Mundell SJ, Bailey CP, Kelly E, Henderson G. Agonist-selective mechanisms of mu-opioid receptor desensitization in human embryonic kidney 293 cells. Mol Pharmacol. 2006;70:676–685. doi: 10.1124/mol.106.022376. [DOI] [PubMed] [Google Scholar]
- 31.Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL, Christopoulos A, Sexton PM, Miller KJ, Spedding M, Mailman RB. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 32.Raehal KM, Schmid CL, Groer CE, Bohn LM. Functional selectivity at the mu-opioid receptor: implications for understanding opioid analgesia and tolerance. Pharmacol Rev. 2011;63:1001–1019. doi: 10.1124/pr.111.004598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pradhan AA, Perroy J, Walwyn WM, Smith ML, Vicente-Sanchez A, Segura L, Bana A, Kieffer BL, Evans CJ. Agonist-Specific Recruitment of Arrestin Isoforms Differentially Modify Delta Opioid Receptor Function. J Neurosci. 2016;36:3541–3551. doi: 10.1523/JNEUROSCI.4124-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhart GD. The Determination of Thermodynamic Allosteric Parameters of an Enzyme Undergoing Steady-State Turnover. Arch Biochem Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 35.Maehle AH, Prull CR, Halliwell RF. The emergence of the drug receptor theory. Nat Rev Drug Discov. 2002;1:637–641. doi: 10.1038/nrd875. [DOI] [PubMed] [Google Scholar]
- 36.Kirk KL. Selective fluorination in drug design and development: an overview of biochemical rationales. Curr Top Med Chem. 2006;6:1447–1456. doi: 10.2174/156802606777951073. [DOI] [PubMed] [Google Scholar]
- 37.Dimagno SG, Sun H. The strength of weak interactions: aromatic fluorine in drug design. Curr Top Med Chem. 2006;6:1473–1482. doi: 10.2174/156802606777951127. [DOI] [PubMed] [Google Scholar]
- 38.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc Natl Acad Sci U S A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith RD, Engdahl AL, Dunbar JB, Jr, Carlson HA. Biophysical limits of protein-ligand binding. J Chem Inf Model. 2012;52:2098–2106. doi: 10.1021/ci200612f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ha JH, Shinsky SA, Loh SN. Stepwise conversion of a binding protein to a fluorescent switch: application to Thermoanaerobacter tengcongensis ribose binding protein. Biochemistry. 2013;52:600–612. doi: 10.1021/bi301105u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tullman J, Nicholes N, Dumont MR, Ribeiro LF, Ostermeier M. Enzymatic protein switches built from paralogous input domains. Biotechnol Bioeng. 2016;113:852–858. doi: 10.1002/bit.25852. [DOI] [PubMed] [Google Scholar]
- 42.Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, McCorvy JD, Jiang Y, Chu MH, Siu FY, Liu W, Xu HE, Cherezov V, Roth BL, Stevens RC. Structural Features for Functional Selectivity at Serotonin Receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang SD, Bruchas MR. Functional Selectivity at GPCRs: New Opportunities in Psychiatric Drug Discovery. Neuropsychopharmacology. 2014;39:248–249. doi: 10.1038/npp.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.