Figure 2. BinA and BinB folds and carbohydrate binding modules.
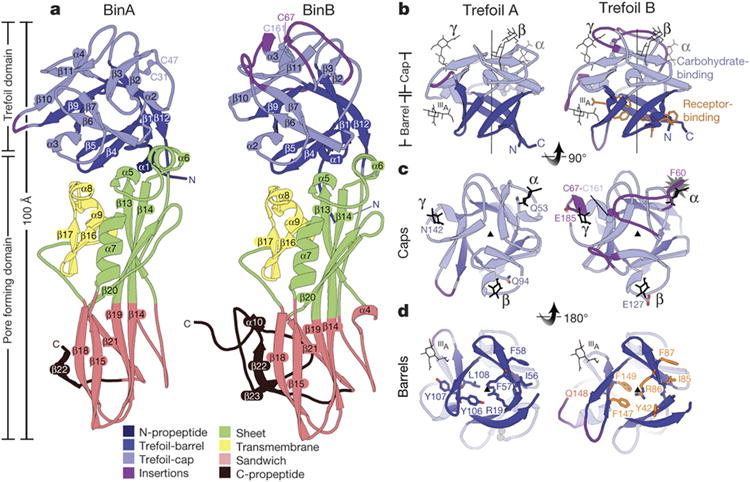
a, BinA and BinB are structurally similar to each other, each composed of trefoil and pore forming domains. The most noticeable differences correspond to insertions in surface loops on the trefoil domains (purple). b, The trefoil domains are composed of barrel and cap subdomains. In the cap subdomain, there are three canonical carbohydrate modules (α, β, γ). Carbohydrate molecules superimposed from structures of hemagglutinin (3AH1)29 and hemolytic lectin (1W3G)30 are shown in black sticks. These occupy the α, β, and γ binding modules. A fourth carbohydrate binding site, marked IIIA, is a minor site observed in hemagglutinin29. c, View of the cap subdomains along the pseudo-three fold symmetry axis; loop insertions (purple) break the symmetry in BinB. The starburst indicates a steric overlap between the modelled carbohydrate and the α module of BinB. The conflict arises from the 9-residue insertion in this loop, tethered by a disulphide bond, C67-C161. d, View of the barrel subdomains along the pseudo three-fold symmetry axis. BinB residues implicated in receptor binding are shown in sticks (orange). Structurally analogous residues are shown on BinA.
