Extended Data Figure 4. Pore-forming domains (PFD) of BinA and BinB.
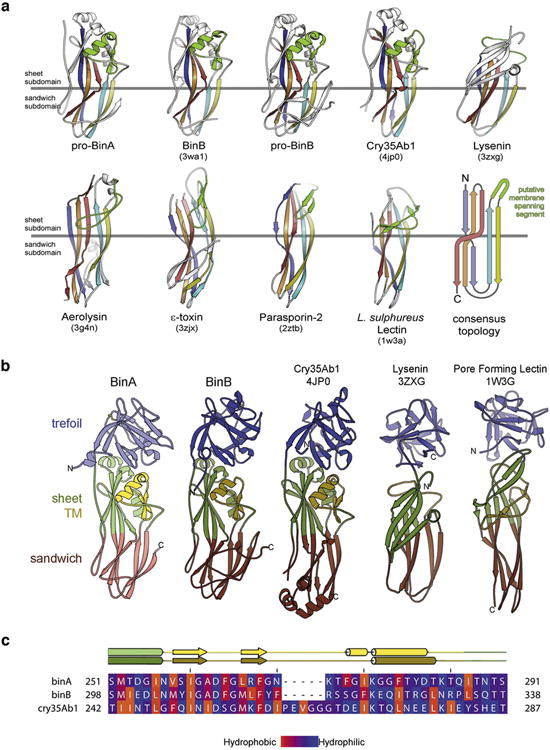
a, Topology of the aerolysin family of pore forming toxins. These share a core topology composed of five antiparallel β-strands and a putative membrane-spanning segment (green). PDB ID codes are included in parentheses. For clarity, we exclude from this illustration any accessory domains outside the pore forming module (PFM) of these toxins. The PFM is divided into two subdomains: a β-sheet subdomain at one end (above the horizontal grey line) and a β-sandwich subdomain at the opposite end (below the horizontal grey line). The length, twist, and number of strands vary between toxins. Also, the putative membrane-spanning segment (green) varies widely in secondary structure. However, in all cases this putative membrane-spanning segment is located between the second and third strands, suggesting these toxins might share a common mechanism of pore formation. b, Members of the aerolysin family that also contain a β-trefoil domain like BinAB. These are: Cry35Ab1 toxin from B. thuringiensis (4jp0) 55, lysenin, a haemolytic toxin from the earthworm Eisenia fetida (3zxg) 56, and a pore-forming lectin from the mushroom Laetiporus suphureus (1w3g) 30. c, Amphipathicity is evident in the sequence of the putative transmembrane spanning subdomain (TM) of BinA and BinB. The observed secondary structures of BinA and BinB are shown above the sequence alignment. The range of the TM subdomain is coloured yellow. Amino acids are coloured by hydrophobicity according to the scale given at the bottom. Note the alternating hydrophobic-hydrophilic pattern is especially prominent in the N-terminal half of the TM subdomain. This pattern is consistent with the proposal of an oligomeric membrane-spanning β-barrel. The figure was made using the program Jalview 57.
