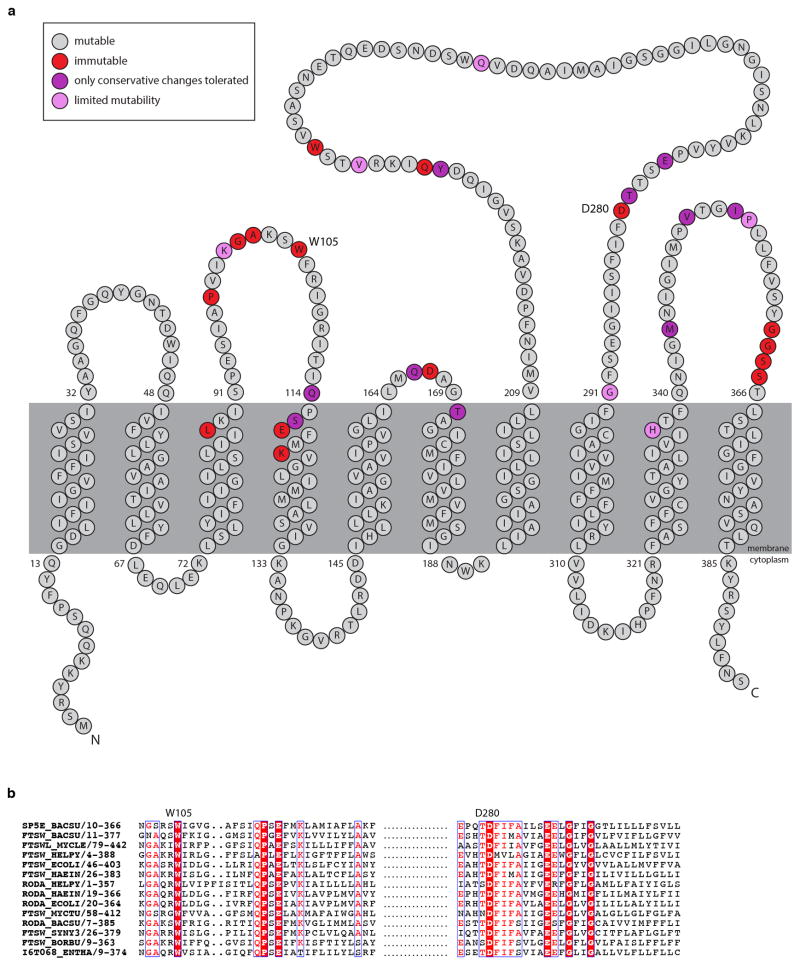
Extended Data Fig. 5. Critical amino acid residues in RodA identified by MutSeq.
a, Topological map of the RodA protein. The extent to which each amino acid residue tolerated mutations based on the MutSeq screen are indicated. Residues that tolerated a spectrum of amino acid changes are shown in grey. Residues that did not tolerate any mutations are shown in red. Residues that only tolerated conservative changes (conservation of charge, hydrophobicity, or functional groups) are in purple. Residues that had limited mutability but tolerated a nonconservative substitution are shown in pink. The complete dataset can be found in Supplementary Table 1. b, Multiple sequence alignment (created using ESPRIPT: http://espript.ibcp.fr/) of 14 diverse SEDS proteins with W105 and D280 residues highlighted.