Figure 3.
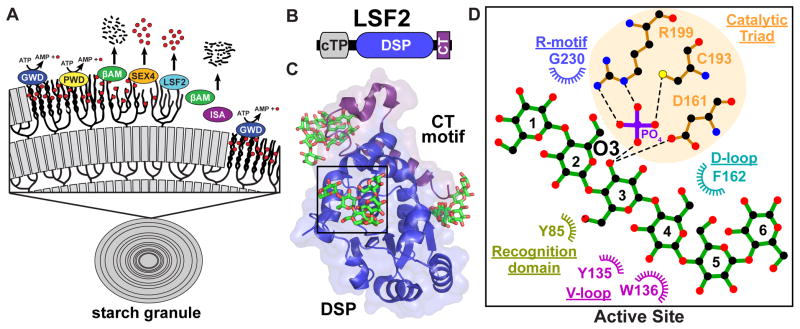
(a) Transitory starch breakdown requires the action of dikinases, phosphatases, and amylases. Glucan, water dikinase (GWD) transfers the β-phosphate of ATP to the C6 position of glucose moieties within the starch lamellae. Phosphoglucan, water dikinase (PWD) then phosphorylates the C3 position of glucose residues. Phosphorylation leads to the disruption of the helices and solubilization of the starch surface, giving β-amylases (βAM) access to degrade the linear chains. SEX4 and LSF2 then remove the C6- and C3-bound phosphate. Dephosphorylation permits β-amylases to continue to degrade the linear chains. Isoamylase (ISA) is required for debranching within the amorphous layer. The cycle resets at the next layer of the starch granule. Red circles represent phosphates. Black dashes and lines represent maltose and glucose. (b) LSF2 is comprised of a chloroplast-targeting peptide (cTP), DSP domain and carboxy-terminal motif (CT), but lacks a CBM. (c) The crystal structure of LSF2 bound to maltohexaose and phosphate (PDB: 4KYR). In addition to phosphate, a single maltohexaose molecule was found at the active site (shown in green). Maltohexaose molecules were also found at two surface binding sites (SBSs) distant from the LSF2 active site. (d) Ligplot+ generated representations of protein-glucan interactions at the LSF2 active site. Glucose moieties are numbered from the non-reducing end to the reducing end. The glucan chain interacts with key residues of the DSP domain with the recognition domain, variable loop (V-loop), D-loop, catalytic triad and R-motif all providing important contacts.