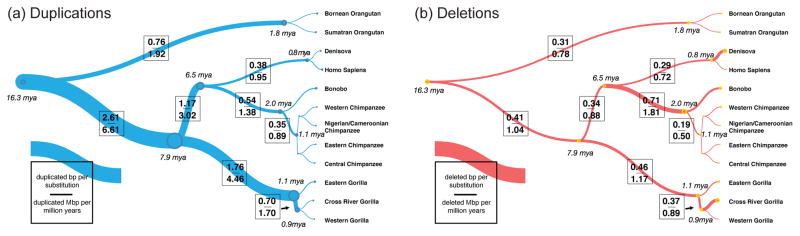
Figure 1. Primate rates of duplication and deletion.
Rates of fixed (a) duplication and (b) deletion are shown as a function of the number of substitutions along each branch of the great ape phylogeny. Branch widths are scaled proportionally to the number of duplicated base pairs per substituted base pair based on analysis of 97 human/ape genomes. A burst of duplicated base pairs appears to have occurred in the common ancestral branch leading to humans and African great apes, where duplicated base pairs were added at 2.6-fold the rate of substitution. In contrast, the rate of deletion in the great ape lineage is more clocklike along all branches (mean of 0.32 deleted base pairs per substitution) with the exception of the chimpanzee–bonobo ancestral lineage, where an approximate twofold increase in the rate of deletion is observed (0.71 deleted base pairs per substitution). Adapted from [10].