Figure 1. Describing the conformational heterogeneity of TFIID.
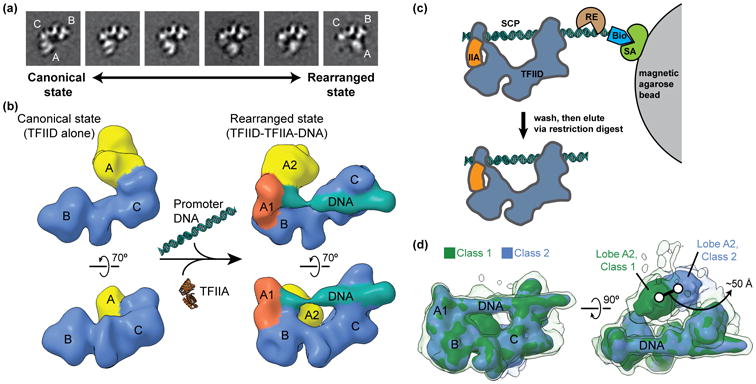
(a) Two-dimensional classification of cryo-EM images of TFIID revealed the conformational rearrangement of TFIID, in which lobe A translocates ∼100 Å across the rigid TFIID core comprised of lobes B and C. Quantification of the occupancy of lobe A in different positions along the BC-core showed that TFIID partitions between the canonical (left) and rearranged (right) states. (b) A multi-model refinement strategy led to 3D reconstructions of TFIID for the both the canonical (left) and rearranged (right) states and showed that TFIID binds to promoter DNA in the rearranged state in the presence of TFIIA (right). (c) Schematic of the strategy used for the purification of promoter-bound TFIID. TFIID was incubated with TFIIA (IIA) and super core promoter (SCP) DNA that was biotinylated (Bio) on one end. The DNA and associated proteins was then immobilized on magnetic streptavidin (SA) coated beads, and excess proteins were washed away. The DNA-bound complexes were then eluted by restriction endonuclease (RE) digest and were subsequently used for the preparation of cryo-EM samples. (d) Characterization of the flexibility of lobe A2 within promoter-bound TFIID. Following alignment to the more rigid promoter-bound BC-core of TFIID (including lobe A1), cryo-EM images of the TFIID-TFIIA-promoter complex were sorted by 3D classification within a mask around the entire range of lobe A. The two major 3D classes are shown in green (class 1) and blue (class 2), with the position of lobe A differing by about 50 Å. Surfaces are displayed at two different intensity thresholds, so that the lower intensity surface in transparency show more flexible elements.
