Figure 2. Improving the resolution of the promoter-bound TFIID core.
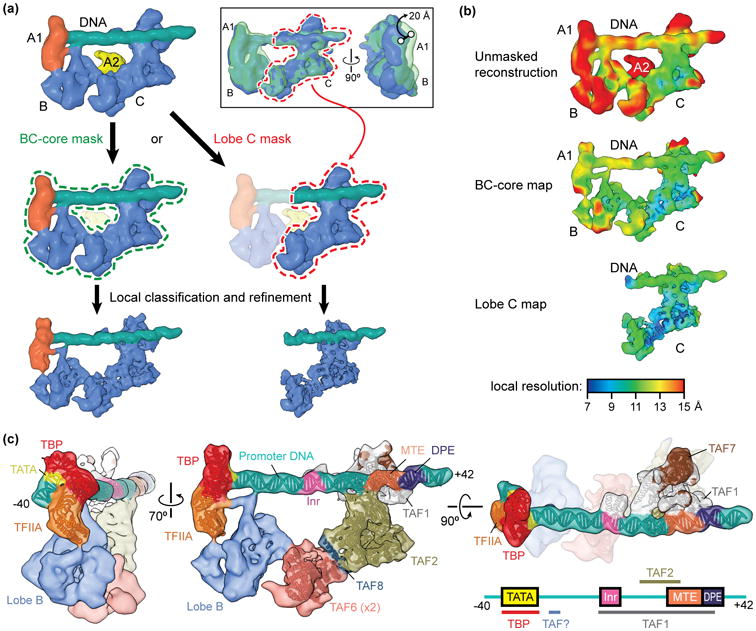
(a) Schematic of focused classification and refinement used to improve the resolution of the promoter-bound BC-core (left) and lobe C (right) of TFIID. Following alignment to the unmasked structure, cryo-EM images of the TFIID-TFIIA-promoter complex were aligned to references with a mask around the entire BC-core or around lobe C only. Images were then sorted by 3D classification within similar masks to extract the best quality particle images within that region. Final refinements of the structures within the masks resulted in improved reconstructions. The inset at the top right shows the flexibility of the lobe B/A1 region of the complex relative to lobe C, as revealed by 3D classification within this region following alignment of the cryo-EM images to lobe C. (b) Local resolution estimation of the unmasked structure (top) versus the BC-core (middle) or lobe C (bottom) reconstructions shows the improvement in resolution obtained by following the focused classification and refinement strategy outlined in (a). (c) Assignments and models of protein subunits and DNA elements within the promoter-bound TFIID structure [24]. The DNA-interacting elements are highlighted in the structure shown on the right, and a diagram of the super core promoter DNA construct used in this study, which contained the TATA, Inr, MTE, and DPE consensus sequences, is shown in the bottom right with the footprints of TFIID subunits indicated, (the identity of one of the DNA-binding TAFs, here labeled “TAF?”, is as of yet unknown).
