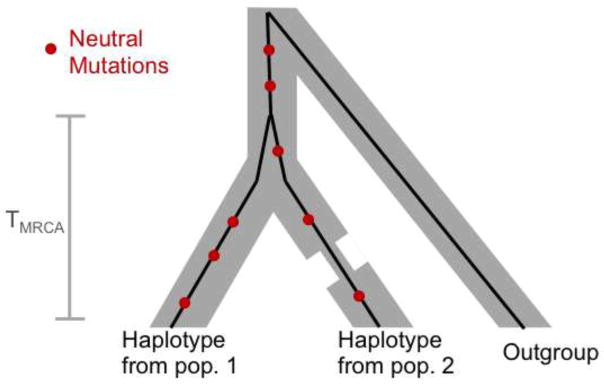
Figure 2.
The genealogy at a locus of one sample from each of two populations, illustrating that the expected number of derived neutral alleles on each sample is the same, and depends only on the time to the most recent common ancestor (TMRCA) of the two lineages and not on the demographic history of the populations.