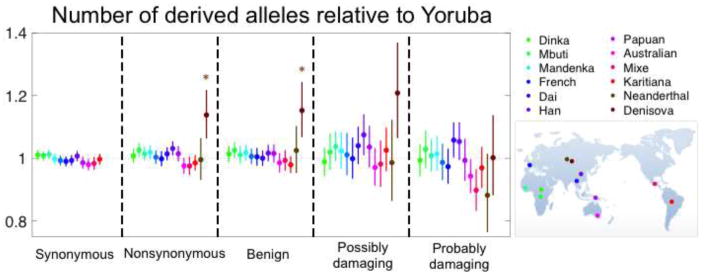
Figure 3.
No significant difference between human populations in the mean number of derived alleles per individual. Each population sample was compared with the Yoruba sample, using data from [32]. Nonsynonymous SNPs were classified into benign, possibly and probably damaging using PolyPhen 2.0 as described in [32]. The numbers of derived alleles for each comparison were counted at sites that were segregating in the joint sample from the two populations, and significance and the shown 95% confidence intervals were evaluated as described in Box 1, dividing the genome into 1,000 blocks. While there are multiple tests performed, it is not obvious how to correct for them, because population samples are also not independent. However, if we assume that at minimum 6 tests were performed then none of the comparisons among human populations is significant at the 5% level. The comparisons with Neanderthal and Denisova incorporate the modifications described in [32], where Denisova are significantly different regardless of the correction for multiple testing.